More Information
Submitted: 18 April 2020 | Approved: 27 April 2020 | Published: 28 April 2020
How to cite this article: Hayashi T, Ura T, Abiko K, Mandan M, Yaegashi N, et al. Reasons why new coronavirus, SARS-CoV-2 infections are likely to spread. J Genet Med Gene Ther. 2020; 3: 001-003.
DOI: 10.29328/journal.jgmgt.1001005
Copyright License: © 2020 Hayashi T, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: ACE2; Furin; SARS-CoV-2; COVID-19; SARS-CoV
Reasons why new coronavirus, SARS-CoV-2 infections are likely to spread
Takuma Hayashi1*, Takashi Ura1, Kaoru Abiko1, Masaki Mandan2, Nobuo Yaegashi3 and Ikuo Konishi1,2,4
1National Hospital Organization Kyoto Medical Center, Kyoto, Kyoto, Japan
2Department of Obstetrics and Gynecology, Kyoto University School of Medicine, Kyoto, Japan
3Department of Obstetrics and Gynecology, Tohoku University School of Medicine, Miyagi, Japan
4Immediate Past President of Asian Society of Gynecologic Oncology, Tokyo, Japan
*Address for Correspondence: Takuma Hayashi D.M.Sci., GMRC, MS, National Hospital Organization, Kyoto Medical Center, Fukakusa Mukai-Cho, Fushimi-Ku, Kyoto-city, Kyoto, Japan, Tel: 81-263-37-2624; Email: [email protected]
The ongoing outbreak of Coronavirus Disease 2019 (COVID-19) originally emerged in China during December 2019 and had become a global pandemic by March 2020. COVID-19 is caused by a novel coronavirus, severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2). Two other coronaviruses have caused world-wide outbreaks in the past two decades, namely SARS-CoV (2002–2003) and Middle East respiratory syndrome coronavirus (MERS-CoV) (2012–present). The surface spike glycoprotein (S), which is critical for virus entry through engaging the host receptor and mediating virus host membrane fusion, is the major antigen of coronaviruses. Recent studies provide molecular insights into antibody recognition of SARS-CoV-2. In this review, we discuss the relationship between the spike glycoprotein of SARS-CoV-2 and its receptor, angiotensin converting enzyme 2 (ACE2) including the latest findings.
Worldwide, the number of patients with new coronavirus infections disease 2019 (COVID-19) have exceeded 200,000. Now, medical researchers are working to find out why the COVID-19 was so widespread. A novel human coronavirus that is now named severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) (formerly called 2019-nCoV) emerged in Wuhan, China, in late 2019 and is now causing a pandemic [1]. The genome of SARS-CoV-2 shares about 80% identity with that of SARS-CoV and is about 96% identical to the bat coronavirus strain BatCoV RaTG13. The SARS-CoV-2 that causes COVID-19 infects human cells much more easily than SARS-CoV [2]. The results of several genome and structural analyzes have revealed that the three dementia structure of SARS-CoV-2, specifically the spike glycoprotein on the virus surface, has characteristics that explain the reason.
The reason that the virus that causes COVID-19 is more infectious than closely related viruses may be due to microscopic features. A team of structural biologist Jason McLellan at the University of Texas at Austin (USA) has analyzed the structure of SARS-CoV-2 by cryo-electron microscopy. That the spike glycoprotein of SARS-CoV-2 binds to ACE2 in human cells, they showed that the binding was at least 10 times stronger than the spike glycoprotein of common SARS coronavirus [3]. The structural information of the spike glycoprotein on surface of SARS-CoV-2 is expected to be useful for vaccine design and therapeutic development for COVID-19.
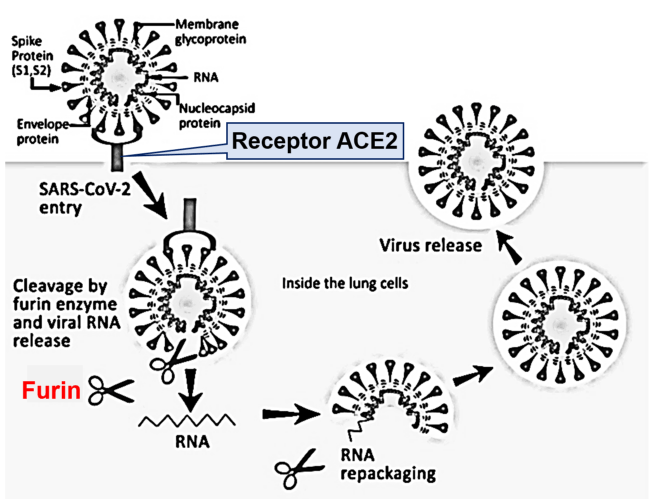
Many researchers have analyzed the receptor on human cell membranes as a gateway for SARS-CoV-2 to enter human tissues. As the first step in infecting cells with SARS-CoV-2, spike glycoproteins on the surface of SARS-CoV-2 bind to cell membranes via ACE2. Genomic analysis of SARS-CoV-2 revealed that the three-dimensional structure of the spike glycoprotein was different from that of closely related viruses. Furthermore, this glycoprotein was shown to have an activation site by the furin enzyme [4] (Figure 1).
Figure 1: Diagram of a coronavirus life cycle. Diagram of a coronavirus life cycle and the various host cell proteases known to cleave and activate some coronavirus Spike glycoproteins. Note that for certain coronaviruses, fusion can occur directly at the plasma membrane. In some producer cell types, SARS-CoV-2 spike proteins are cleaved by furin/proprotein convertases in the exocytic pathway. Cleaved SARS-CoV-2 spike proteins change their conformations rapidly after receptor binding, exposing subsequent proteolytic cleavage sites, which are processed by proteases (i.e., TMPRs, found at or near cell surfaces).
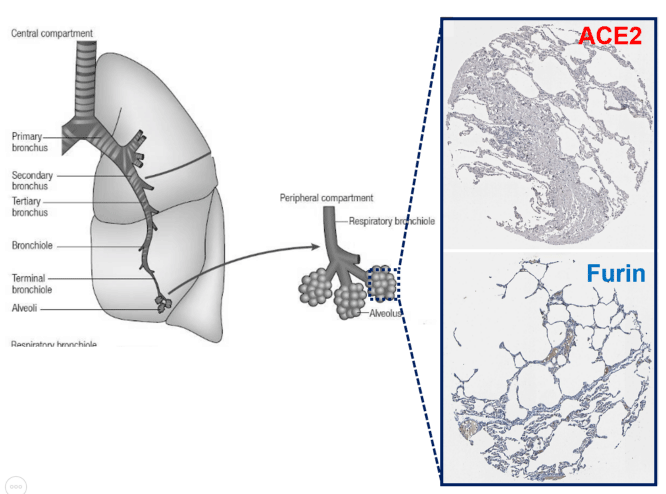
Furin is an enzyme in the proportion convertase family which cleaves precursor proteins and facilitates their conversion to a biologically active state. Furin is a member of the subtilisin-like proportion convertase family, which includes proteases that process protein and peptide precursors trafficking through multiple branches of the secretory pathway. It is a type 1 membrane-bound protease that is expressed in multiple tissues. Furin is indispensable in maintaining peripheral tolerance, which is due, at least in part, to its no redundant essential functioning in regulating transforming growth factor-β1 (TGF-β1) production [5]. Furin is expressed in significant concentrations in the lungs (Figure 2). Thus, coronaviruses in the respiratory tract can make use of this enzyme to convert and activate their own surface glycoproteins [6,7]. This makes their role in viral protein processing noteworthy.
Figure 2: Expressions of ACE2 and Furin in the alveolar epithelial cells. The major function of the lungs is respiratory exchange, and inhaled air is conducted to the alveoli (the small sacs in which gaseous exchange occurs) through a network of branching tubes (bronchi and bronchioles). The larger tubes, having muscular walls reinforced with cartilage are the bronchi, while smaller tubes (< 1 mm in diameter) that have incomplete muscular walls and lack cartilage are called bronchioles, and they connect to the alveoli. Immunohistochemistry expression of ACE2 and Furin in the normal alveoli epithelial cells.
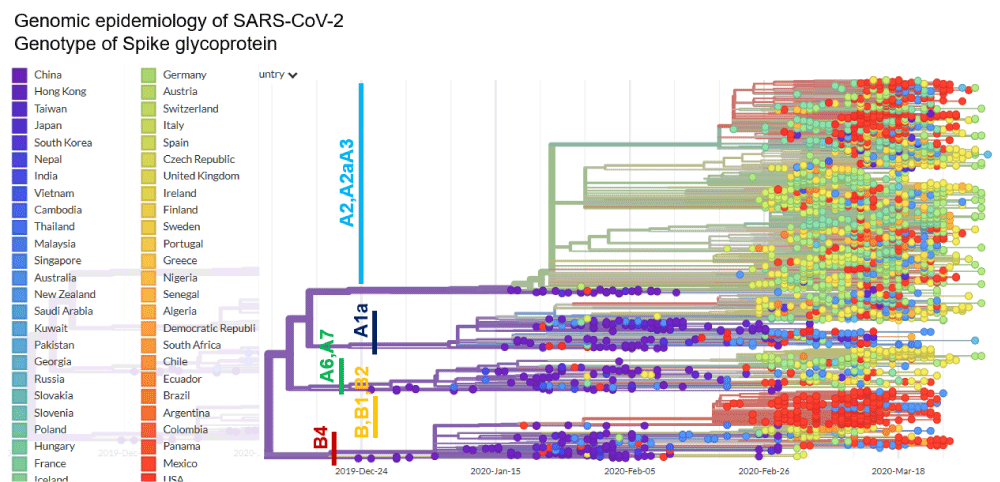
As a result of furin’s ability to cleave important cell surface proteins, it and other proprotein convertases are the target of considerable research interest. Frequently the spike glycoprotein is cleaved by a host protease (furin-like protease) into two functional domains, S1 and S2 [8]. S1 mainly helps in receptor binding, while S2 gives structural support in the form of the stalk of Spike glycoprotein on surface of coronavirus [8]. Furin inhibitors can be targeted as potential drug therapies for SARS-CoV-2. The S1/S2 cleavage site is the target for furin during infection [7]. It is important to note that other beta coronaviruses do not contain this cleavage site. SARS-CoV, which is closely related to SARS-CoV-2, does not bear a cleavage site. Some of the most pathogenic forms of influenza have similar cleavage sites, which can be acted upon by furin and other cellular proteases. In a phylogenetic network analysis of amino acid sequences of spike glycoprotein derived from 160 complete human SARS-Cov-2 genomes, we find ten central variants distinguished by amino acid changes, which we have named A1a, A2, A2a, A3, A6, A7, B, B1, B2, and B4, with being the ancestral type according to the bat outgroup coronavirus. The A1a, A6, A7 and B4 types are found in significant proportions in East Asia, A2, A2a, and A3 types are found in Europeans, B, B1, and B2 types are found in Americans. In contrast, the B4 type is the most common type in America and East Asia (Figure 3). The phylogenetic classification provided here may be used to rule out or confirm such effects when evaluating clinical and epidemiological outcomes of SARS-CoV-2 infection, and when designing treatment and, eventually, vaccines.
Figure 3: Phylogenetic network analysis of amino acid sequences of spike glycoprotein derived from 160 complete human SARS-Cov-2 genomes. Our research results find ten central variants distinguished by amino acid changes, which we have named A1a, A2, A2a, A3, A6, A7, B, B1, B2, and B4, with being the ancestral type according to the bat out group coronavirus. The A1a, A6, A7 and B4 types are found in significant proportions in East Asia, A2, A2a, and A3 types are found in Europeans, B, B1, and B2 types are found in Americans. In contrast, the B4 type is the most common type in America and East Asia.
The ubiquitous expression of cellular proteases across cell types increases the potential for the virus to successfully infiltrate the host. Any organs such as human lung, liver and small intestine. The distribution of the expression of furin means that SARS-CoV-2 can attack many organs. Symptoms such as liver failure found in some infected individuals may be due to furin activity. Recent research findings provide a structural framework to identify conserved and accessible epitopes across spike glycoproteins that will support ongoing vaccine design efforts. Finally, elicitation of diverse, polyclonal antibody responses might prove key role in light of the diversity of viruses circulating in animal reservoirs and in preventing the possible emergence of SARS-CoV-2 neutralization escape mutants.
It points out that the furin activation site may facilitate SARS-CoV-2 human-to-human transmission.
Health officials say the virus is transported only through droplets that are coughed or sneezed out - either directly, or on objects. But some scientists say there is preliminary evidence that airborne transmission - in which the disease spreads in the much smaller particles from exhaled air, known as aerosols - is occurring, and that precautions, such as increasing ventilation indoors, should be recommended to reduce the risk of infection [9,10]. Regarding the new coronavirus, SRAS-CoV-2, the World Health Organization (WHO) reports that about two-thirds of all deaths in Europe are men. There are still many unknowns about this virus, and it is necessary to conduct research from a technical point of view.
Footnote
The protease furin is induced on T cell activation and is a target of signal transducers and activators of transcription (STAT)-transcription factors in T cells. Studying furin has been hampered by the fact that germline deletion of this gene is embryonically lethal. Furin’s physiological role in T cells was studied using a T cell-specific furin conditional knockout mice. The striking finding is that conditional deletion of furin in T cells results in loss of peripheral tolerance and systemic autoimmune disease [8]. Furin deficiency compromises TGF-β1 release and regulatory T cell function, and is associated with inherently more aggressive effector T cells. Recent research finding suggests that inhibiting furin may promote the immune response but may also cause the loss of peripheral tolerance by reducing levels of bioavailable TGF-β1 [8].
Data sharing
Data are available on various websites and have been made publicly available (more information can be found in the first paragraph of the Results section).
Disclosure of potential conflicts of interest
The authors declare no potential conflicts of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We sincerely thank Professor Richard A. Young (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology) for his research assistance. This study was supported in part by grants from the Japan Ministry of Education, Culture, Science and Technology (No. 24592510, No. 15K1079, and No. 19K09840), The Foundation of Osaka Cancer Research, The Ichiro Kanehara Foundation for the Promotion of Medical Science and Medical Care, The Foundation for the Promotion of Cancer Research, The Kanzawa Medical Research Foundation, The Shinshu Medical Foundation, and The Takeda Foundation for Medical Science.
Author contributions
T.H. performed most of the experiments and coordinated the project; T.H. and M.M. conceived the study and wrote the manuscript. N.Y. and I.K. gave information on clinical medicine and oversaw the entire study.
- Coronavirus disease (COVID-2019) situation reports. Geneva: World Health Organization. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579: 270-273. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/32015507
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367: 1260-1263. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/32075877
- Andersen KG, Rambaut A, Lipkin WI, Edward C, Holmes EC, et al. The proximal origin of SARS-CoV-2. Nature Medicine. 2020; 26: 450–452.
- Pesu M, Watford WT, Wei L, Xu L, Fuss I, et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008; 455: 246-250. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/18701887
- Ji HL, Zhao R, Matalon S, Matthay MA. Elevated Plasmin(ogen) as a Common Risk Factor for COVID-19 Susceptibility. Physiol Rev. 2020; 100: 1065-1075. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/32216698
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020: 181: 281-292. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/32155444
- Abraham S, Kienzle TE, Lapps W, Brian DA. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990; 176: 296-301. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/2184576
- Shi J, Wen Z, Zhong G, Yang H, Wang C, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020; pii: eabb7015. Pubmed: https://www.ncbi.nlm.nih.gov/pubmed/32269068
- Lewis d. Is the coronavirus airborne? Experts can’t agree. Nature. 2020.