More Information
Submitted: September 16, 2022 | Approved: September 28, 2022 | Published: September 29, 2022
How to cite this article: Antoniadis P, Gheorghe FA, Nitu MA, Nitu CG, Constantinescu DR, et al. New insights of liquid biopsy in ovarian cancer. J Genet Med Gene Ther. 2022; 5: 001-011.
DOI: 10.29328/journal.jgmgt.1001007
Copyright License: © 2022 Antoniadis P, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Liquid biopsy; CTCs; miRNA; cfDNA; Ovarian cancer
New insights of liquid biopsy in ovarian cancer
Panagiotis Antoniadis1*, Florentina Alina Gheorghe2, Madalina Ana Maria Nitu3, Cezara Gabriela Nitu4, Diana Roxana Constantinescu5 and Florentina Duica6*
1GenDx & GenDx Products, Yaelaan 48, 3584 CM Utrech, The Netherlands
2Nova Group Investment, Street Narciselor 22, Dragomirești-Vale, Ilfov, 077095, Romania
3Sanador, Street Dr. Iacob Felix 32, Bucharest, 011038, Romania
4Elias Emergency University Hospital, Bulevardul Mărăști 17, Bucharest, 011461, Romania
5Centrul Medical Baneasa, Strada Neagoe Vodă 3-5, Bucharest, 077190, Romania
6Bucharest Emergency Clinical Hospital, Calea Floreasca 8, Bucharest, 014461, Romania
*Address for Correspondence: Panagiotis Antoniadis, GenDx & GenDx Products, Yaelaan 48, 3584 CM Utrech, The Netherlands, Email: [email protected]
*Address for Correspondence: Florentina Duica, Bucharest Emergency Clinical Hospital, Calea Floreasca 8, Bucharest, 014461, Romania, Email: [email protected]; [email protected]
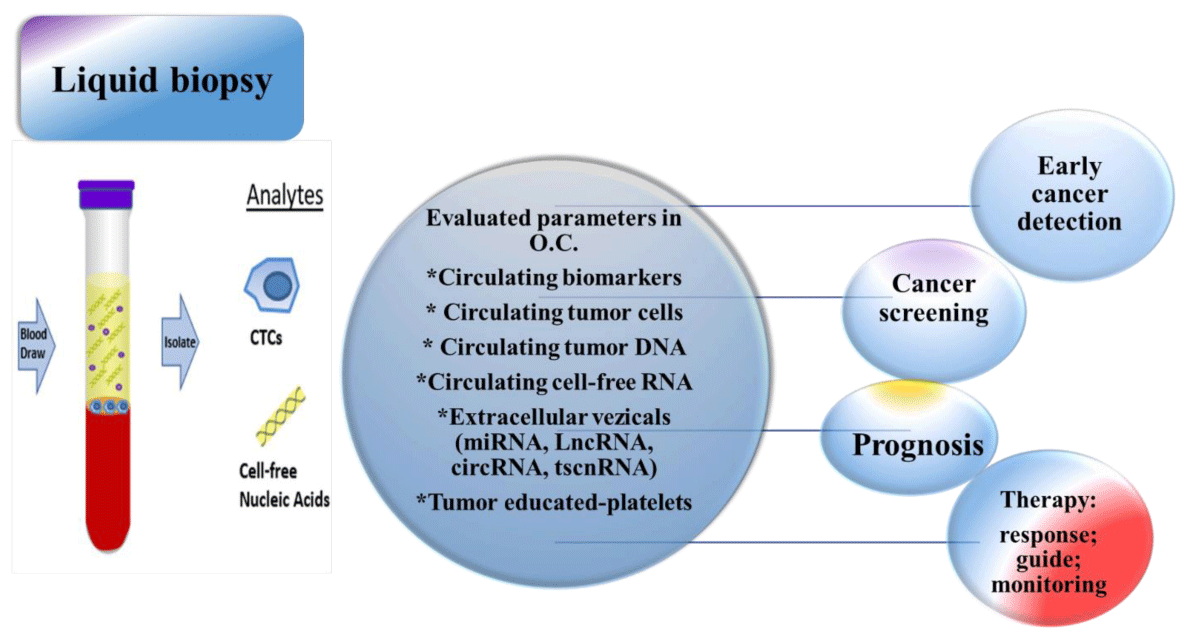
Through the development of new analysis technologies, many issues regarding the approach to tumoral diseases have been elucidated. With analytical assays developed in the last years, various omics technologies have evolved in such a manner that the characteristics of tumor cells and products can be evaluated (assessed) in the bloodstream of cancer patients at different times. Ovarian Cancer (OC) is one of the most difficult to diagnose umors, with low survival rates due to the high heterogeneity of these diseases that are distinct in terms of etiology and molecular characteristics, but which simply share an anatomical appearance. Recent findings have indicated that several types of ovarian cancer classified into different histotypes are in fact derived from non-ovarian issues and share few molecular similarities. Within this context, ovarian cancer screening and diagnosis can be made through the evaluation of circulating tumor cells in peripheral blood using liquid biopsy technologies. Advances in the study of various molecules analyzed by liquid biopsy have shown that elucidation of intratumoural and intertumoural heterogeneity and spatial and temporal tumor evolution could be traced by serial blood tests rather than by histopathological analyses of tissue samples from a primary tumor. Therefore, evaluation of some molecules such as circulating tumor cells (CTC), circulating tumor DNA (ctDNA), circulating cell-free RNA (non-coding and mRNA, extracellular vesicles), tumor-educated platelets or different miRNAs using liquid biopsy could lead to improvement of patient management.
OC: Ovarian Cancer; TVUS: Transvaginal Ultrasound; CT: Computerized Tomography; PET-CT: Positron Emission Computerized Tomography; HE4: Human Epididymis Protein 4; ISLB: International Society of Liquid Biopsy; ICGC: International Cancer Genome Consortium; FDA - Food Drug Administration; TCGA: The Cancer Genome Atlas; HTAN: Human Tumor Atlas Network; ILSA: International Liquid Biopsy Standardization Alliance; FNIH: The Foundation for the National Institutes of Health; BC: Biomarkers Consortium; QCMs: Quality Control Materials; ENCODE: Encyclopedia Of DNA Elements; BEAM: Beads, Emulsion, Amplification, Magnetics; cfDNA: Circulating cell-free DNA; cfRNA: Circulating cell-free RNA; CTC: Circulating Tumor Cells; ctDNA: Circulating tumor DNA; circRNA: Circular RNA; HE4: Human Epididymis Protein 4; LncRNA: Long non-coding RNA; TsncRNAs: Trans- noncoding RNA; miRNA: Micro RNA; mRNA: messenger RNA; NGS: Next Generation Sequencing; OC: Ovarian Cancer; PCR: Polymerase Chain Reaction; WGS: Whole-Genome Sequencing; WES: Whole-Exome Sequencing
The aim of this study is to provide an overview of recent findings in the field of diagnosis and evaluation of prognosis, including the possibility of detection of small residual tumors of ovarian carcinomas, in order to improve the management of particular cases. In this article, we summarized the main used biomarkers in OC diagnosis that could be analyzed through liquid biopsy assays, which is a non-invasive technique that allows serial sampling collection and analysis, for monitoring dynamic tumor changes over time. Liquid biopsy as a tool for diagnosing cancer refers to a simple and less invasive procedure that could provide valuable information in clinical oncology just by analyzing a small number of tissues, often a single peripheral blood sample.
Ovarian cancer has the highest mortality rate of all gynecological cancers worldwide being frequently diagnosed at an advanced stage [1,2].
The classification of ovarian cancer is generally made according to the stage at the time of tumor discovery, early or advanced stage and according to the histology of the tumor, classified as epithelial or non-epithelial, of which the most frequent is the high-grade serous ovarian carcinoma (HGSOC) [2-4]. This type of cancer is characterized by an unusual dissemination mechanism: rapid growth, disruption of ovarian tumor capsules and malignant cells spread into the peritoneal cavity which usually involves the accumulation of ascites. Therefore, the development of new methods for investigating circulating cells is widely reported as a rational avenue for improving clinical efficiency and preventing the progression of disease in patients with ovarian cancer. Given the challenges of screening, diagnosis and monitoring, and the impact that the early diagnosis has on the patient’s prognosis, new emerging tools are needed to increase the precision of the diagnosis and to monitor and better understand and predict the response to the treatment [4,5].
In the context of ovarian cancers, classical diagnostic and screening procedures are not efficient, leading to late-stage detection of the disease, which results in inefficient treatments and poor survival rates. Usually, tumors of the ovary are diagnosed by conventional methods such as tissue biopsy, imaging techniques (transvaginal ultrasound - TVUS), computerized tomography scan (CT, PET-CT) and evaluation of some biomarkers from circulating blood. Ovarian tissue biopsy is obtained by different invasive techniques such as surgical, needle biopsy, or imaging-guided-biopsy, in which some solid tissue fragments are removed for pathological examination. In these procedures, limitations of collecting the right pieces of the samples can restrict the information about the tumor heterogeneity and detection of possible metastasis in other sites. As for biomarkers, they can be divided into several categories, such as biomarkers specific for the diagnosis of OC (one of the commonly used combinations being the CA125, Human Epididymis Protein 4 - HE4 and mesothelin), biomarkers used as a prognosis for OC among which we could list predictive biomarkers and treatment response biomarkers, more recently used in personalized medicine in patients with OC [6,7]. Over the last decade, many studies have been made for the detection and management of OC and numerous molecules have been considered promising diagnostic and/or prognostic biomarkers, such as CTC and ctDNA. In this context, we aim to discover the newest and relevant research in the domain of OC and we carried out a literature search in public databases such as PubMed, Google Academic, NCBI, Science Direct, etc., using keywords liquid biopsy and ovarian cancer.
Liquid biopsy - non-invasive molecular analysis tool used in diagnosis and therapies ajustement in OC
Liquid biopsy is a technique that refers to the evaluation of some types of cells or sub-cellular structures from biological fluids, by diverse molecular analysis. Due to the promising potential of this non-invasive method of analysis, liquid biopsy is used nowadays in clinical practice in prenatal screening and in special in oncology for investigating circulating cells, sub-cellular structures, or other bio-molecules. These types of molecules are considered biomarkers that fulfill important roles in diagnostic, prognostic or monitoring therapy response, in order to change the treatment in a timely manner to prevent the recurrence of the disease [8,9]. Several single and multi-gene assays using different omics technologies for detecting genetic alteration of some circulating cells that carried out genetic information about molecular profiling of the primary, metastatic or recurrent tumors were approved by international organizations. International Cancer Genome Consortium (ICGC), Food Drug Administration (FDA), The Cancer Genome Atlas (TCGA), Human Tumor Atlas Network and the International Liquid Biopsy Standardization Alliance (ILSA) has made large collaborative studies, with the purpose of better understanding tumor biology and elucidating the intratumoral and intertumoral heterogeneity. Other processes intensively studied lately are represented Through the interaction between tumoral cells, metabolites that these cells release in the microenvironmental space and their interaction with the genome. Furthermore, the mechanisms by which metabolic and homeostatic processes are dysregulated and monitoring therapy response or targeted therapy have evolved. This progress was made in order to apply standardization of some biomarkers that could be used for screening to provide chances of early detection of cancers or for monitoring the response to targeted therapy.
Use of liquid biopsy in clinical practice
As a result of numerous technological advances in the fields of genetics, genomics, and oncology, promising developments in precision medicine have evolved lately. Among these signs of progress, liquid biopsy ranks one of the first places in customization of individual cases, by evolution made in the approach regarding diagnostic and therapy. Today, liquid biopsy is used as an alternative to solid biopsies, due to the non-invasive characteristic of this procedure, by which genetic material necessary to evaluate the genomic variation of a tumor, is obtained. By liquid biopsy, some types of molecules are obtained in order to characterize or evaluate the prognostic and evolution of the tumor. These include CTC and ctDNA, being the most used biomolecules to customize health care for individual patients [10-13]. In this scope, the International Society of Liquid Biopsy (ISLB), founded in 2017, deals with the regulation and standardization of recommendations in the field of diagnosis by liquid biopsy. The main purpose of this society is to coordinate the research efforts and strategies of all specialists worldwide. Few working groups have put their efforts together to introduce in clinical practice the use of many biomarkers obtained and quantified by liquid biopsy. One of them is The Foundation for the National Institutes of Health (FNIH), Biomarkers Consortium (BC), whose main the research area is represented by biomarker use, especially in new drug development and medical diagnostics in cancers patients and discovery and implementation of Quality Control Materials (QCMs) like in the ctDNA Quality Control Materials Project [10,15-17]. The importance of liquid biopsy in clinical practice it`s recognized in oncology by supporting the clinical decision in order to classify and characterize cancer types, by identifying the mutation for targeted therapy, monitoring therapy response [10,15], as it is shown in Figure 1.
Figure 1: The importance of liquid biopsy in clinical practice.
In ovarian cancer, although many biomarkers are available in the clinic, does not exist yet a set of specific universally accepted and used biomarkers that allow early diagnosis and monitoring disease progression. In the following chapters, we will discuss the most commonly used types of biomolecules in ovarian cancer, obtained through liquid biopsy.
Circulating biomarkers in OC usually detected through liquid biopsy
Many molecules produced by tumoral cells or cells in the environment of a cancer type could be evaluated in body fluids, especially in peripheral blood, by qualitative or quantitative techniques. These molecules can be considered a tumor biomarkers, because they originate from various cellular sources such as cytoplasmic proteins, surface antigens or receptors, enzymes, hormones, or another specific type cells, especially oncogenes and their products [14]. Various isolation methods have been developed either based on biological or by physical properties for CTC, ctDNA, circulating cell-free RNA (non-coding and messenger RNA, extracellular vesicles), tumor-educated platelets, or different miRNAs, which are evaluated by liquid biopsy. CTCs are detected by several methods of gene expression analysis (RT-PCR, FISH), by immunocytochemistry (ICC) or a combination of both [15].
Circulating tumor cells (CTC): CTCs are major components of liquid biopsies for clinical diagnosis, prognosis and real-time treatment monitoring [18,19], as their significance was first reported by Thomas Ashworth in 1869 [20]. These cancer cells preserve characteristics and the heterogeneity of the primary tumor, while they can potentially initiate subsequent metastases. Due to their significance, several technologies have been developed, targeting the enrichment, isolation, and detection of CTCs in liquid biopsies. In total, these technologies can be categorized into 3 groups: the ones that probe the biological [20,21] or physical [23,24] properties of CTCs and the functional assays [25,26], as summarized in Table 1.
| Table 1: Technologies for the isolation and detection of CTCs in liquid biopsies. | ||
| Targeting method | Targets | References |
| Biological properties of CTCs. Positive and/or negative selection. |
Positive selection: EpCAM, cytokeratins such as CK18, CK19 and CK8. Negative selection: CD45, CD66b antigens. |
[27,28] |
| Physical properties of CTCs |
size, density, electrostatic properties and the greater ability of CTCs to deform. |
[29] |
| Functional assays | Protein secretion from CTCs, preferential binding of CTCs to | [30,31] |
The prognostic value of CTCs is focused on the ability to discriminate potentially metastatic clones, while the elucidation of the molecular and biological properties of CTCs can facilitate optimized treatment. Monitoring of transcripts in CTCs have been shown to have substantial predictive value for prostate cancer patients [32], while probing for CTCs in HR-positive breast cancer patients has been shown to have the potency to predict late recurrence [33]. Several studies have denoted the significance of molecular and biological characteristics of CTCs in the early differential diagnosis of cancer [34] and prevention of metastasis [35]. Continuous enumeration of CTCs in cancer patients has been shown to be a valuable tool for prognosis and treatment evaluation [36,37].
Circulating tumor DNA (ctDNA): Apart from CTCs, the thought of investigating liquid biopsies has been mainly based on ctDNA, as it is an accessible target that facilitates prognosis and treatment monitoring, while it has the potential to become an early cancer diagnosis tool. The release of ctDNA into circulation is done by passive mechanisms, such as cell death, as well as active mechanisms of cells, such as the release of extracellular vesicles [38]. Previously identified mutations in primary tumors can be found in ctDNA to monitor the responsiveness of treatment [39], whereas novel lesions can be indicated with a non-targeted sequencing approach [40]. The fact that ctDNA is mixed with cfDNA from healthy cells renders difficult the detection of variants, however, several technologies have demonstrated effective detection of variants having frequency down to 0.01% [41]. The detection of variants can be done among others by digital PCR, NGS, or BEAMing [42]. As cancer development is characterized by epigenetic modifications as well, there is an increasing interest to identify specific aberrant methylation patterns in ctDNA, mainly in promoter regions [43]. Generally, ctDNA has been found to be increased in cancer patients compared to healthy individuals [44], while the prognostic value of ctDNA has been indicated by several studies connecting specific variants [45] or methylation markers [46] with the survival rates of the patients. The assessment of ctDNA has been found to be vital for the selection of therapy in cancer patients with difficult-to-resect tumors [47] and for monitoring treatment resistance [48]. In the study of Willis, et al. microsatellite instability assessment has been performed using ctDNA, which is a key biomarker for the administration of immunotherapies in several cancer types, including ovarian [49].
Circulating cell-free RNA (non-coding and mRNA): The presence of cfRNAs in the blood is associated with the levels they have at the tissues of origin and their release rate from the cells. As in several cancer types is hard to obtain adequate cfDNA from the plasma of the patients, due to low concentrations, cfRNA could fill in the gap and facilitate early cancer type and subtype diagnosis together with localization of the primary tumor. The significance of cfRNA as a diagnostic, prognostic, and treatment response biomarker has been mainly studied focusing on miRNAs and mRNAs (messenger RNA). Although it has been shown that changes in the expression levels of specific miRNAs could be used in cancer monitoring [50], assays targeting only miRNAs lack reproducibility and specificity as they are prone to processing biases [51]. The investigation of mRNAs with known cancer-driving mutations or fusions and the assessment of their expression levels has substantial usage in clinical practice [52]. The selection of specific mRNAs that are not found in the plasma of healthy individuals but are present in the liquid biopsies of cancer patients could supplement cfDNA assays in cancer monitoring [53]. Targeting biomarkers that are tissue-specific, such as overexpressing cfRNAs or methylation patterns on cfDNA, could facilitate clinical practice in the prediction of tumor tissues of origin (TOO).
Extracellular vesicles: There are three types of EVs that can be differentiated by their size and biogenesis, namely exosomes, microvesicles and apoptotic bodies [54]. The smallest ones, ranging from 30 to 150 nm in diameter, are the exosomes, which are currently considered an important component of cell-to-cell communication [55]. Their cargo contains target molecules for cancer monitoring, such as miRNA, mRNA and cfDNA, they could facilitate clinical practice by providing a reflection of the cancer cells they originate from. The stress conditions which are exerted on cancer cells can alter normal vesicular biogenesis and exocytosis together with the composition of the vesicles. Exosomes can be found in adequate concentrations in liquid biopsies, while they are very stable and tolerant to transportation [56,57]. These attributes together with the fact that the target molecules, such as sensitive cfRNA, remain protected within their lipid bilayer render them a promising source of biomarkers.
Tumor-educated platelets (TEPs): Among others, the functionality of platelets has been associated with thrombus formation to induce hemostasis, the communication of immune cells, the propagation of inflammation, as well as the creation of new blood vessels. It has been noted that there is an active interplay between cancer cells and TEPs which helps metastasis from intravasation to extravasation and the formation of metastatic niche, through epithelial to mesenchymal transition (EMT) induction of cancer cells, intravascular protection of cancer cells in clots and neoangiogenesis at the metastatic site [58-60]. The “education” process of platelets takes place through the direct acquisition of biomolecules from the tumor and the cells around it via extracellular vesicles, indirectly through post-transcriptional splicing of RNAs in platelets as well as through altering the transcriptional profile of megakaryocytes [61,62]. Even though platelets lack a nucleus, they are capable of protein synthesis, which gives them an active role in cancer propagation [63]. Considering that TEPs contain molecules of interest, such as RNAs from cancer cells, could be targeted by assays to contribute to early cancer diagnosis, prognosis and treatment response prediction [64].
Use of biomarkers obtained through liquid biopsy as a prognostic tool in personalized medicine in OC patients
The conventional biomarkers used in the clinical diagnosis of ovarian cancer are serum cancer antigen 125 (CA125), Human Epididymis Protein 4 (HE4) and transvaginal ultrasonography. Using these biomarkers alone for differential diagnosis is insufficient due to the low sensitivity and specificity of these procedures, so, many studies were made and furthermore are coming up in order to explore the relation between miRNA and ovarian cancer and to improve diagnosis, prognosis and treatment methods. The development of tumors is imposed by the tumor microenvironment. Extracellular matrix molecules regulate cancer invasion and metastasis and, at the same time, downregulation of miRNAs controls tumor spreading by degrading extracellular matrix [65,66].
miRNAs are used as a source of liquid biopsy in ovarian cancer: One of the RNA types involved in cancer oncogenesis, prognosis, and treatment response is microRNA (miRNA), a small, non-coding RNA, containing between 19-25 nucleotides, with multiple roles in almost all cell functions, such as cell differentiation, proliferation, or death. The miRNA is endogenously synthesized in the cell nucleus, by DNA polymerase 2 and its precursor is processed by Drosha and Pasha enzymes and transported into the cytoplasm. The 70-100 nucleotides pre-miRNA are then cleaved by Dicer into a functional 22 nucleotides miRNA [67,68]. The miRNA regulates the post-transcriptional gene expression by getting incorporated in an mRNA silencing complex, by inhibiting the mRNA translation or by degrading the mRNA. This process is based on its complementarity with the target zone, although it is not always necessary, as one miRNA can sometimes regulate more than one mRNA [67,69,70]. Oncogenesis can be influenced by both the downregulation and upregulation of miRNA. By downregulating miRNA, the tumor genesis can be suppressed, whereas upregulation of miRNA can act as an enhancer, promoting abnormal cell growth dysregulating apoptosis, promoting neovascularization and an inflammatory environment [71]. Because of its multiple roles in cell biology, miRNA can also be used as a biomarker in medical practice, as it is expressed in fluids like blood and its components plasma and serum, in urine, saliva and breast milk, in both oncologic patients and healthy people. The genetic signature of circulating miRNA is identical to the tissular miRNA, thus providing more accessible tools for detecting the presence of ovarian cancer, its histological type, prognosis, and [71,72]. MiRNA can be used as a biomarker due to its stability in the bloodstream, even in extreme conditions such as variations in pH or temperature. Its degradation is prevented by binding to serum proteins or lipids or by being incorporated into small vesicles that resulted from apoptosis [73,74]. Dosing the circulating miRNA can be used as a diagnostic tool, as some specific miRNAs are modified in ovarian cancer patients compared to healthy individuals. In a study from 2006, Zhang, et al. found that the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141) is upregulated in ovarian cancer, while a study in 2015 showed that miR- 92, miR-15a, and miR-21 are also upregulated in patients with ovarian cancer [75,76]. Some miRNA may also be associated with certain ovarian cancer histology. The miR-200 family is found in all subtypes of ovarian cancer, but endometrioid tumors presented upregulated miR-21 and downregulated miR-222. Other miRNAs are downregulated in ovarian cancer patients, for example, miR-9, miR-31, miR-34, miR-503, miR-506 and miR-507 [ 67-69,100]. The miRNA can also be used as a prognostic tool. For example, as found in a study from 2016 by Nakamura, patients with lower levels of miR-200 family or with lower methylation of let 7a-3, had a lower survival rate [77]. Another study group demonstrated that miR-21 can be used as an independent prognostic factor in ovarian cancer, as its higher values are associated with more advanced disease, lower histologic differentiation, and lower survival [74]. Another role of miRNA is to predict the response to therapy, as indicated in a study by Kapetanakis, et al. They showed that the levels of miR-200b during chemotherapy can be correlated to the treatment response. For patients with decreasing levels of miR-200b, the progression-free survival was longer than in those with increasing levels of miR-200b, who had an increased risk of progression [78]. Oncologic treatment was developed by making use of the miRNA and its function, the treatment of cachexia associated with inflammation and high levels of miR-21 being one such example. Inhibition of microvesicles containing miR-21 and their interaction with myocytes has been a promising strategy for the treatment of severe weight and muscle loss in cancer patients [73]. By studying miRNAs in ovarian cancer patients, we have gained insights into the complexity of their roles in cellular mechanisms, in both normal and cancerous cells. Their clinical utility is yet to be determined, but recent studies have given promising perspectives on using them in daily practice as easy-to-determine biomarkers [73]. In Table 2 we have summarized the role of some miRNA associated with ovarian carcinoma.
| Table 2: Role of miRNAs in OC. | ||
| Structure | Role of miRNAs in OC | References |
| miR-21-5p | miR21 transferred from CAAs to the cancer cells confer chemoresistance by binding to its direct novel target- APAF1 | [73,79] |
| miR-506 | miR506 downregulation promotes an aggressive phenotype in OC | [100] |
| miR-141 | Is a b-Catenin, TCF7L2, SOX17 inhibitor/inhibitor/activator Up-modulated in ovarian carcinoma | [69] |
| miR-126 | miR126 may serve tumor suppressor roles by inducing G1 cell cycle arrest and suppressing invasion in ovarian cancer cells, by targeting VEGF expression | [101] |
| miR-200 family | EMT regulation (double-negative feedback loop) Up-regulated in ovarian tumors compared to normal cells and tissues | [75-78,84] |
LncRNAs, used as a source of liquid biopsy in ovarian cancer: Recently, a significant role in ncogenesis has been attributed to the long non-coding RNAs (lncRNAs), from an oncogenic perspective, and more scientists, are studying this subject for a better understanding of its impact in the field of cancer, especially ovarian cancer [86]. LncRNAs are a subclass of non-coding RNAs, that is not translated into protein and that contains approximately 200 nucleotides, which differs from short ncRNAs [87]. They have a big impact on the cell environment as transcriptional regulators, due to the specific interactions with other cell components, such as proteins, DNA, or RNA [88]. Mutations in the structure of lncRNA can change the basic cell functions, leading to the growth of cancer tumors. Therefore, lncRNA could be used as a useful biomarker in the development of cancer, such as ovarian cancer [86]. In ovarian cancer development, lncRNA acts as a process suppressor, as it can induce autophagy in the cell. A remarkable example is lncRNA GAS8-AS1, which demonstrated this capability by binding to the multi-domain protein Beclin1, the main compound in the process of autophagy. Also, Meg3, a lncRNA that has a specific role in ovarian oncogenesis, suppressing tumoral transformation, has been identified by both Oliveira, et al. and Zamaraev, et al. [86,89]. Regarding the impact on cell apoptosis, it has been observed that the apoptotic process is affected by lncRNA, which acts on the tumor suppressor p53, by regulating it. Chemoresistance in patients with OC can be induced when lncRNA suppresses p53 protein or Bcl-2 family proteins (proteins with pro-apoptotic action) [87]. Several studies have shown that lncRNA is involved in fundamental signaling pathways, such as PI3K/Akt signaling, affecting the process of malignant proliferation [86]. In the immune system, the lncRNA is involved in the release of an anti-tumor immune response, which leads to OC cells being undetected by immune system cells, making lncRNA a goal in OC immunotherapy [90]. One way to find drugs for OC patients could be to use compounds that could act directly on the lncRNA. Thus, by means of lncRNA, it may be possible to regulate the apoptosis of OC cells [91]. Nevertheless, not all the actions and functions of lncRNA is yet known. Several studies in this area will help researchers in the future to lay the groundwork for ovarian cancer therapy, targeting the lncRNA directly [92].
circRNAs in OC: Circular RNAs are RNA molecules that belong to the lncRNA class and they range in size from hundreds of nucleotides to thousands of nucleotides [93]. Their structure is in the form of a closed, circular loop and they are not usually translated into proteins and have a non-polyadenylated structure [94]. The important characteristics of circular RNA are represented by the ubiquitous presence, the stability of their structure, their conserved structure, but also the multitude of roles that they fulfill, such as the involvement in the process of splicing and translation regulation. The circular shape of the molecules generates a much more stable structure against RNases, compared to the linear structure of other RNA molecules [95]. Over time, scientists have shown major involvement of circular RNA molecules in most cancers, including OC. The action of circular RNAs in OC is to control the process of cell proliferation. It has been observed that there is a close association between the stages of initiation and progression of ovarian cancer and the deregulation of circular RNA molecules, which leads to the use of circular RNA as biomarkers [93]. Ning, et al. found that circular RNA has basic functions in oncogenesis because it regulates the expression of certain genes. Thus, they found that 6 types of circular RNA (circ-EXOC6B, circ-BNC2, circ-FAM13B, circ- N4BP2L2, circCELSR1 and circ-RHOBTB3) are associated with certain clinical and pathological features of ovarian cancer. They also found that the circular RNA molecules, circEXOC6B and circ- N4BP2L2, can be used as biomarkers for the prognosis of ovarian cancer [96].
tsncRNA used as a source of liquid biopsy in ovarian cancer: TsncRNAs (trans- noncoding RNA) belong to small non-coding RNA, a relatively new described class of bio-molecules that are involved in various biological processes, such as regulation of gene expression at transcriptional and posttranscriptional levels [97]. Like it was demonstrated by ENCODE project (Consortium, E.P. The ENCODE (Encyclopedia of DNA Elements), 90% of the whole human genome contains functional ncRNA [98]. In the last decade, it was well documented that EVs isolated and analyzed by liquid biopsies, are a carrier of various cargo, including tsncRNA molecules, and serve to transport genetic material between cells, with involvement in, regulating important biological processes, such as the proliferation and differentiation of cells [99]. Recent studies have shown that circulating tsncRNA molecules are present in 14 biological samples processed in patients diagnosed with epithelial ovarian cancer or other ovarian tumors, thus suggesting their potential as biomarkers for both diagnosis and prognosis.
Main difficulties of biomarker research in ovarian carcinomas
Despite the rapid progress in the field of liquid biopsy technology, however, there is still a gap between research studies realized on cohorts of patients with ovarian cancers and clinical practice application of these findings.
Liquid biopsy is not a routinely used diagnostic test, due to the lack of uniform screening strategies between countries across the world. Although these procedures, could obtain valuable information about the quick identification of the clonal evolution of cancer cells and also monitorisation of the development of changes leading to drug resistance in the future, there are limitations in collecting, separating and analyzing the right biomarkers in the blood sample due to many altered cells involved in the carcinogenic process. Another concern raised by the application of liquid biopsy to cancer screening in the general population is made by false-positive results that could be obtained, because of the constant accumulation of genetic and epigenetic alteration in the microenvironment of the tumoral site and free circulation of diverse molecules that are evaluated through this technique [105,106].
Liquid biopsy assays are obtained by evaluation of large gene panels through whole-genome sequencing (WGS) or whole-exome sequencing (WES) data analysis of tumor tissue samples and also could be customized into personalized gene panel, but the effectiveness and potential economic impact of including this procedure into general health screening programs are not yet justified. For that, the public healthcare systems must establish collaboration with multiple stakeholders such as patient organizations, the regulatory and policymakers authorities to support economic cost and to establish the conditions of implementation and the medical and scientific community.
The development of multi-analyte panels correlates with analise of patient history, clinical data and liquid biopsy marker profiles are essential to improve diagnostic accuracy [102-104].
Although many biomarkers have been studied over time, and some of them have been used to assess the status, progression and efficacy of the drug therapy in ovarian cancer, till now, is still no clear set of specific biomolecules which can be used as a reference standard in ovarian cancer detection or in evaluating therapy. In the last decade, liquid biopsy has evolved as a revolutionary technique by which divers genomic and proteomic information about patients with cancers can be deciphering. The various biomolecules are separated by the technique of liquid biopsy can be both diagnostic and prognostic biomarkers in OC and can be used to predict tumorigenesis in ovarian cancers and other types of gynecological tumors. With the development of new technologies in the field of molecular biology such as NGS (next generation sequencing), biological samples obtained by liquid biopsy techniques can be processed, which provide data on specific mutations that may occur in target genes involved in cancer processes. This information is extremely useful both for the diagnosis, prognosis and screening of various types of cancer, as well as for predicting a patient’s response or resistance to receiving treatments, which may allow early diagnosis of disease progression.
Author contributions
Conceptualization, F.D., P. A.; methodology, F.D., F.A.G, D. C; investigation, P. A., M. A.M. N., C.G.N, F. A. G., D. R. C. and. F.D.; writing—original draft preparation, P. A. , F. A. G., M. A.M. N., C.G.N, D. R. C., F.D.; writing—review and editing, F. A. G., F.D., M. A.M. N., C.G.N., supervision F.D., P. A.; project administration, F. D., F. A. G., M. A.M. N.
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010 Sep;177(3):1053-64. doi: 10.2353/ajpath.2010.100105. Epub 2010 Jul 22. PMID: 20651229; PMCID: PMC2928939.
- Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005 May;5(5):355-66. doi: 10.1038/nrc1611. PMID: 15864277.
- Cochrane DR, Tessier-Cloutier B, Lawrence KM, Nazeran T, Karnezis AN, Salamanca C, Cheng AS, McAlpine JN, Hoang LN, Gilks CB, Huntsman DG. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin? J Pathol. 2017 Sep;243(1):26-36. doi: 10.1002/path.4934. Epub 2017 Aug 7. PMID: 28678427.
- Idrees R, Din NU, Siddique S, Fatima S, Abdul-Ghafar J, Ahmad Z. Ovarian seromucinous tumors: clinicopathological features of 10 cases with a detailed review of the literature. J Ovarian Res. 2021 Mar 18;14(1):47. doi: 10.1186/s13048-021-00796-y. PMID: 33736662; PMCID: PMC7977580.
- Brown J, Frumovitz M. Mucinous tumors of the ovary: current thoughts on diagnosis and management. Curr Oncol Rep. 2014 Jun;16(6):389. doi: 10.1007/s11912-014-0389-x. PMID: 24777667; PMCID: PMC4261626.
- Chandra A, Pius C, Nabeel M, Nair M, Vishwanatha JK, Ahmad S, Basha R. Ovarian cancer: Current status and strategies for improving therapeutic outcomes. Cancer Med. 2019 Nov;8(16):7018-7031. doi: 10.1002/cam4.2560. Epub 2019 Sep 27. PMID: 31560828; PMCID: PMC6853829.
- Macías M, Alegre E, Díaz-Lagares A, Patiño A, Pérez-Gracia JL, Sanmamed M, López-López R, Varo N, González A. Liquid Biopsy: From Basic Research to Clinical Practice. Adv Clin Chem. 2018;83:73-119. doi: 10.1016/bs.acc.2017.10.003. Epub 2017 Nov 23. PMID: 29304904.
- Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020 Jan 1;468:59-71. doi: 10.1016/j.canlet.2019.10.014. Epub 2019 Oct 11. PMID: 31610267.
- Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020 Jan 1;468:59-71. doi: 10.1016/j.canlet.2019.10.014. Epub 2019 Oct 11. PMID: 31610267.
- Connors D, Allen J, Alvarez JD, Boyle J, Cristofanilli M, Hiller C, Keating S, Kelloff G, Leiman L, McCormack R, Merino D, Morgan E, Pantel K, Rolfo C, Serrano MJ, Pia Sanzone A, Schlange T, Sigman C, Stewart M. International liquid biopsy standardization alliance white paper. Crit Rev Oncol Hematol. 2020 Dec;156:103112. doi: 10.1016/j.critrevonc.2020.103112. Epub 2020 Sep 30. PMID: 33035734.
- Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019 Oct;19(10):553-567. doi: 10.1038/s41568-019-0180-2. Epub 2019 Aug 27. PMID: 31455893.
- Chang L, Ni J, Zhu Y, Pang B, Graham P, Zhang H, Li Y. Liquid biopsy in ovarian cancer: recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics. 2019 May 31;9(14):4130-4140. doi: 10.7150/thno.34692. PMID: 31281536; PMCID: PMC6592165.
- Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019 Mar;205:77-91. doi: 10.1016/j.trsl.2018.10.003. Epub 2018 Oct 12. PMID: 30391474.
- Omer Devaja and Andreas Papadopoulos, INTECHOPEN LIMITED, Ovarian cancer- from pathogenesis to treatment; 2018; http://dx.doi.org/105772/66599
- Asante DB, Calapre L, Ziman M, Meniawy TM, Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020 Jan 1;468:59-71. doi: 10.1016/j.canlet.2019.10.014. Epub 2019 Oct 11. PMID: 31610267.https://doi.org/10.1016/j.canlet.2019.10.014
- Anfossi S. Clinical utility of circulating non-coding RNAs – an update. Nat. Rev. Clin. Oncol. 15 September (9), 2018;541–563. https://doi.org/10.1038/s41571- 018-0035-x
- 2022March2nd;Availablefrom:https://www.nibsc.org/science_and_research/advanced_therapies/genomic_reference_materials.aspx.
- Soler A, Cayrefourcq L, Mazard T, Babayan A, Lamy PJ, Assou S, Assenat E, Pantel K, Alix-Panabières C. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci Rep. 2018 Oct 29;8(1):15931. doi: 10.1038/s41598-018-34365-z. PMID: 30374140; PMCID: PMC6206091.
- Poudineh M, Sargent EH, Pantel K, Kelley SO. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat Biomed Eng. 2018 Feb;2(2):72-84. doi: 10.1038/s41551-018-0190-5. Epub 2018 Feb 6. PMID: 31015625.
- Ashworth TR. A Case of Cancer in Which Cells Similar to Those in the Tumours Were Seen in the Blood after Death. The Medical Journal of Australia. 1869; 14:146-147.
- Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, Fritsche HA, Hortobagyi GN, Terstappen LW. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005 Mar 1;23(7):1420-30. doi: 10.1200/JCO.2005.08.140. Erratum in: J Clin Oncol. 2005 Jul 20;23(21):4808. PMID: 15735118.
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007 Dec 20;450(7173):1235-9. doi: 10.1038/nature06385. PMID: 18097410; PMCID: PMC3090667.
- Antfolk M, Magnusson C, Augustsson P, Lilja H, Laurell T. Acoustofluidic, label-free separation and simultaneous concentration of rare tumor cells from white blood cells. Anal Chem. 2015 Sep 15;87(18):9322-8. doi: 10.1021/acs.analchem.5b02023. Epub 2015 Sep 3. PMID: 26309066.
- Abdulla A, Liu W, Gholamipour-Shirazi A, Sun J, Ding X. High-Throughput Isolation of Circulating Tumor Cells Using Cascaded Inertial Focusing Microfluidic Channel. Anal Chem. 2018 Apr 3;90(7):4397-4405. doi: 10.1021/acs.analchem.7b04210. Epub 2018 Mar 20. PMID: 29537252.
- Tulley S, Zhao Q, Dong H, Pearl ML, Chen WT. Vita-Assay™ Method of Enrichment and Identification of Circulating Cancer Cells/Circulating Tumor Cells (CTCs). Methods Mol Biol. 2016;1406:107-19. doi: 10.1007/978-1-4939-3444-7_9. PMID: 26820949.
- Togo S, Katagiri N, Namba Y, Tulafu M, Nagahama K, Kadoya K, Takamochi K, Oh S, Suzuki K, Sakurai F, Mizuguchi H, Urata Y, Takahashi K. Sensitive detection of viable circulating tumor cells using a novel conditionally telomerase-selective replicating adenovirus in non-small cell lung cancer patients. Oncotarget. 2017 May 23;8(21):34884-34895. doi: 10.18632/oncotarget.16818. PMID: 28432274; PMCID: PMC5471019.
- de Wit S, van Dalum G, Terstappen LW. Detection of circulating tumor cells. Scientifica (Cairo). 2014;2014:819362. doi: 10.1155/2014/819362. Epub 2014 Jul 15. PMID: 25133014; PMCID: PMC4124199.
- Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014 Mar;9(3):694-710. doi: 10.1038/nprot.2014.044. Epub 2014 Feb 27. PMID: 24577360; PMCID: PMC4179254.
- Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016 Mar;10(3):374-94. doi: 10.1016/j.molonc.2016.01.007. Epub 2016 Jan 28. PMID: 26897752; PMCID: PMC5528969.
- Friedlander TW, Ngo VT, Dong H, Premasekharan G, Weinberg V, Doty S, Zhao Q, Gilbert EG, Ryan CJ, Chen WT, Paris PL. Detection and characterization of invasive circulating tumor cells derived from men with metastatic castration-resistant prostate cancer. Int J Cancer. 2014 May 15;134(10):2284-93. doi: 10.1002/ijc.28561. Epub 2014 Jan 2. PMID: 24166007.
- Ramirez JM, Fehm T, Orsini M, Cayrefourcq L, Maudelonde T, Pantel K, Alix-Panabières C. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem. 2014 Jan;60(1):214-21. doi: 10.1373/clinchem.2013.215079. Epub 2013 Nov 19. PMID: 24255082.
- Miyamoto DT, Lee RJ, Kalinich M, LiCausi JA, Zheng Y, et al. An RNA-Based Digital Circulating Tumor Cell Signature Is Predictive of Drug Response and Early Dissemination in Prostate Cancer. Cancer Discov. 2018 Mar;8(3):288-303. doi: 10.1158/2159-8290.CD-16-1406. Epub 2018 Jan 4. PMID: 29301747; PMCID: PMC6342192.
- Sparano J, O'Neill A, Alpaugh K, Wolff AC, Northfelt DW, Dang CT, Sledge GW, Miller KD. Association of Circulating Tumor Cells With Late Recurrence of Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018 Dec 1;4(12):1700-1706. doi: 10.1001/jamaoncol.2018.2574. PMID: 30054636; PMCID: PMC6385891.
- Guo W, Sun YF, Shen MN, Ma XL, Wu J, et al. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin Cancer Res. 2018 May 1;24(9):2203-2213. doi: 10.1158/1078-0432.CCR-17-1753. Epub 2018 Jan 26. PMID: 29374055.
- Zhou Q, Geng Q, Wang L, Huang J, Liao M, Li Y, Ding Z, Yang S, Zhao H, Shen Q, Pan C, Lou J, Lu S, Chen C, Luo Q. Value of folate receptor-positive circulating tumour cells in the clinical management of indeterminate lung nodules: A non-invasive biomarker for predicting malignancy and tumour invasiveness. EBioMedicine. 2019 Mar;41:236-243. doi: 10.1016/j.ebiom.2019.02.028. Epub 2019 Mar 12. PMID: 30872130; PMCID: PMC6442989.
- Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, Peng W, Sandhu SK, Olmos D, Riisnaes R, McCormack R, Burzykowski T, Kheoh T, Fleisher M, Buyse M, de Bono JS. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015 Apr 20;33(12):1348-55. doi: 10.1200/JCO.2014.55.3487. Epub 2015 Mar 23. PMID: 25800753; PMCID: PMC4397279.
- Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y, Cao Y, Shen L. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer. 2016 Jan 19;114(2):138-45. doi: 10.1038/bjc.2015.417. PMID: 26784122; PMCID: PMC4815805.
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013 Aug;10(8):472-84. doi: 10.1038/nrclinonc.2013.110. Epub 2013 Jul 9. PMID: 23836314.
- Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015 Jan;61(1):112-23. doi: 10.1373/clinchem.2014.222679. Epub 2014 Nov 11. PMID: 25388429.
- Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res. 2016 Dec 1;22(23):5772-5782. doi: 10.1158/1078-0432.CCR-16-1231. Epub 2016 Sep 6. PMID: 27601595; PMCID: PMC5448134.
- Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016 Jul 26;7(30):48832-48841. doi: 10.18632/oncotarget.9453. PMID: 27223063; PMCID: PMC5217053.
- Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault SF. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. 2018 Jan;18(1):7-17. doi: 10.1080/14737159.2018.1400384. Epub 2017 Nov 13. PMID: 29115895.
- Nunes SP, Diniz F, Moreira-Barbosa C, Constâncio V, Silva AV, Oliveira J, Soares M, Paulino S, Cunha AL, Rodrigues J, Antunes L, Henrique R, Jerónimo C. Subtyping Lung Cancer Using DNA Methylation in Liquid Biopsies. J Clin Med. 2019 Sep 19;8(9):1500. doi: 10.3390/jcm8091500. PMID: 31546933; PMCID: PMC6780554.
- Alix-Panabières C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med. 2012;63:199-215. doi: 10.1146/annurev-med-062310-094219. Epub 2011 Nov 2. PMID: 22053740.
- Cheng H, Liu C, Jiang J, Luo G, Lu Y, Jin K, Guo M, Zhang Z, Xu J, Liu L, Ni Q, Yu X. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int J Cancer. 2017 May 15;140(10):2344-2350. doi: 10.1002/ijc.30650. Epub 2017 Mar 9. PMID: 28205231.
- Xu RH, Wei W, Krawczyk M, Wang W, Luo H, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017 Nov;16(11):1155-1161. doi: 10.1038/nmat4997. Epub 2017 Oct 9. PMID: 29035356.
- Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018 Nov 1;379(18):1754-1765. doi: 10.1056/NEJMra1706174. PMID: 30380390.
- Del Re M, Crucitta S, Gianfilippo G, Passaro A, Petrini I, Restante G, Michelucci A, Fogli S, de Marinis F, Porta C, Chella A, Danesi R. Understanding the Mechanisms of Resistance in EGFR-Positive NSCLC: From Tissue to Liquid Biopsy to Guide Treatment Strategy. Int J Mol Sci. 2019 Aug 14;20(16):3951. doi: 10.3390/ijms20163951. PMID: 31416192; PMCID: PMC6720634.
- Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin Cancer Res. 2019 Dec 1;25(23):7035-7045. doi: 10.1158/1078-0432.CCR-19-1324. Epub 2019 Aug 4. PMID: 31383735.
- Cheung KWE, Choi SR, Lee LTC, Lee NLE, Tsang HF, Cheng YT, Cho WCS, Wong EYL, Wong SCC. The potential of circulating cell free RNA as a biomarker in cancer. Expert Rev Mol Diagn. 2019 Jul;19(7):579-590. doi: 10.1080/14737159.2019.1633307. Epub 2019 Jun 24. PMID: 31215265.
- Lee I, Baxter D, Lee MY, Scherler K, Wang K. The Importance of Standardization on Analyzing Circulating RNA. Mol Diagn Ther. 2017 Jun;21(3):259-268. doi: 10.1007/s40291-016-0251-y. PMID: 28039578; PMCID: PMC5426982.
- Yeung DT, Hughes TP. Therapeutic targeting of BCR-ABL: prognostic markers of response and resistance mechanism in chronic myeloid leukaemia. Crit Rev Oncog. 2012;17(1):17-30. doi: 10.1615/critrevoncog.v17.i1.30. PMID: 22471662.
- Larson MH, Pan W, Kim HJ, Mauntz RE, Stuart SM, Pimentel M, Zhou Y, Knudsgaard P, Demas V, Aravanis AM, Jamshidi A. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun. 2021 Apr 21;12(1):2357. doi: 10.1038/s41467-021-22444-1. Erratum in: Nat Commun. 2022 May 4;13(1):2553. PMID: 33883548; PMCID: PMC8060291.
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018 Nov 23;7(1):1535750. doi: 10.1080/20013078.2018.1535750. PMID: 30637094; PMCID: PMC6322352.
- Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017 Jan 26;7(3):789-804. doi: 10.7150/thno.18133. PMID: 28255367; PMCID: PMC5327650.
- Hu T, Wolfram J, Srivastava S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer. 2021 Feb;7(2):122-133. doi: 10.1016/j.trecan.2020.09.003. Epub 2020 Sep 30. PMID: 33008796.
- Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y, Zheng S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020 Aug 3;5(1):144. doi: 10.1038/s41392-020-00258-9. PMID: 32747657; PMCID: PMC7400738.
- Schlesinger M. Role of platelets and platelet receptors in cancer metastasis. J Hematol Oncol. 2018 Oct 11;11(1):125. doi: 10.1186/s13045-018-0669-2. PMID: 30305116; PMCID: PMC6180572.
- Asghar S, Parvaiz F, Manzoor S. Multifaceted role of cancer educated platelets in survival of cancer cells. Thromb Res. 2019 May;177:42-50. doi: 10.1016/j.thromres.2019.02.026. Epub 2019 Feb 26. PMID: 30849514.
- Buergy D, Wenz F, Groden C, Brockmann MA. Tumor-platelet interaction in solid tumors. Int J Cancer. 2012 Jun 15;130(12):2747-60. doi: 10.1002/ijc.27441. Epub 2012 Feb 28. PMID: 22261860.
- In 't Veld SGJG, Wurdinger T. Tumor-educated platelets. Blood. 2019 May 30;133(22):2359-2364. doi: 10.1182/blood-2018-12-852830. Epub 2019 Mar 4. PMID: 30833413.
- Best MG, Wesseling P, Wurdinger T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018 Jul 1;78(13):3407-3412. doi: 10.1158/0008-5472.CAN-18-0887. Epub 2018 Jun 19. PMID: 29921699.
- Zhang Q, Liu H, Zhu Q, Zhan P, Zhu S, Zhang J, Lv T, Song Y. Patterns and functional implications of platelets upon tumor "education". Int J Biochem Cell Biol. 2017 Sep;90:68-80. doi: 10.1016/j.biocel.2017.07.018. Epub 2017 Jul 25. PMID: 28754316.
- Tjon-Kon-Fat LA, Sol N, Wurdinger T, Nilsson RJA. Platelet RNA in Cancer Diagnostics. Semin Thromb Hemost. 2018 Mar;44(2):135-141. doi: 10.1055/s-0037-1606182. Epub 2017 Sep 13. PMID: 28905353.
- Cochrane DR, Tessier-Cloutier B, Lawrence KM, Nazeran T, Karnezis AN, Salamanca C, Cheng AS, McAlpine JN, Hoang LN, Gilks CB, Huntsman DG. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin? J Pathol. 2017 Sep;243(1):26-36. doi: 10.1002/path.4934. Epub 2017 Aug 7. PMID: 28678427.
- Idrees R, Din NU, Siddique S, Fatima S, Abdul-Ghafar J, Ahmad Z. Ovarian seromucinous tumors: clinicopathological features of 10 cases with a detailed review of the literature. J Ovarian Res. 2021 Mar 18;14(1):47. doi: 10.1186/s13048-021-00796-y. PMID: 33736662; PMCID: PMC7977580.
- Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: An overview. J Cell Physiol. 2018 May;233(5):3846-3854. doi: 10.1002/jcp.26095. Epub 2017 Aug 25. PMID: 28703277.
- Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009 Jul 3;284(27):17897-901. doi: 10.1074/jbc.R900012200. Epub 2009 Apr 1. PMID: 19342379; PMCID: PMC2709356.
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007 Sep 15;67(18):8699-707. doi: 10.1158/0008-5472.CAN-07-1936. PMID: 17875710.
- Prahm KP, Høgdall C, Karlsen MA, Christensen IJ, Novotny GW, Knudsen S, Hansen A, Jensen PB, Jensen T, Mirza MR, Ekmann-Gade AW, Nedergaard L, Høgdall E. Clinical validation of chemotherapy predictors developed on global microRNA expression in the NCI60 cell line panel tested in ovarian cancer. PLoS One. 2017 Mar 23;12(3):e0174300. doi: 10.1371/journal.pone.0174300. PMID: 28334047; PMCID: PMC5363866.
- Gov E, Kori M, Arga KY. Multiomics Analysis of Tumor Microenvironment Reveals Gata2 and miRNA-124-3p as Potential Novel Biomarkers in Ovarian Cancer. OMICS. 2017 Oct;21(10):603-615. doi: 10.1089/omi.2017.0115. Epub 2017 Sep 22. PMID: 28937943.
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008 Jul;110(1):13-21. doi: 10.1016/j.ygyno.2008.04.033. Erratum in: Gynecol Oncol. 2010 Jan;116(1):153. PMID: 18589210.
- Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016 Jun 24;15(1):48. doi: 10.1186/s12943-016-0536-0. PMID: 27343009; PMCID: PMC4921011.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008 Jul 29;105(30):10513-8. doi: 10.1073/pnas.0804549105. Epub 2008 Jul 28. PMID: 18663219; PMCID: PMC2492472.
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008 May 13;105(19):7004-9. doi: 10.1073/pnas.0801615105. Epub 2008 May 5. PMID: 18458333; PMCID: PMC2383982.
- Zhang DZ, Lau KM, Chan ES, Wang G, Szeto CC, Wong K, Choy RK, Ng CF. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PLoS One. 2014 Jul 11;9(7):e100793. doi: 10.1371/journal.pone.0100793. PMID: 25014919; PMCID: PMC4094487.
- Xu C, Zeng Q, Xu W, Jiao L, Chen Y, Zhang Z, Wu C, Jin T, Pan A, Wei R, Yang B, Sun Y. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013 Feb;12(2):207-19. doi: 10.1158/1535-7163.MCT-12-0273. Epub 2012 Dec 27. PMID: 23270926.
- Kapetanakis NI, Uzan C, Jimenez-Pailhes AS, Gouy S, Bentivegna E, Morice P, Caron O, Gourzones-Dmitriev C, Le Teuff G, Busson P. Plasma miR-200b in ovarian carcinoma patients: distinct pattern of pre/post-treatment variation compared to CA-125 and potential for prediction of progression-free survival. Oncotarget. 2015 Nov 3;6(34):36815-24. doi: 10.18632/oncotarget.5766. PMID: 26416421; PMCID: PMC4742212.
- Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, Schmandt R, Lu KH, Mok SC. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016 Mar 29;7:11150. doi: 10.1038/ncomms11150. PMID: 27021436; PMCID: PMC4820618.
- Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y, Rupaimoole R, Rodriguez-Aguayo C, Lopez-Berestein G, Ji P, Chen K, Sood AK, Mezzanzanica D, Liu J, Sun B, Zhang W. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015 Jan;235(1):25-36. doi: 10.1002/path.4443. Epub 2014 Nov 6. PMID: 25230372; PMCID: PMC4268369.
- Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer. 2015 Aug;51(12):1638-49. doi: 10.1016/j.ejca.2015.04.021. Epub 2015 May 26. PMID: 26025765.
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007 Sep 15;67(18):8699-707. doi: 10.1158/0008-5472.CAN-07-1936. PMID: 17875710.
- Rhodes LV, Martin EC, Segar HC, Miller DF, Buechlein A, Rusch DB, Nephew KP, Burow ME, Collins-Burow BM. Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015 Jun 30;6(18):16638-52. doi: 10.18632/oncotarget.3184. PMID: 26062653; PMCID: PMC4599295.
- Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, Urban N, Knudsen BS, Tewari M. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010 Jan;116(1):117-25. doi: 10.1016/j.ygyno.2009.08.009. Epub 2009 Oct 24. PMID: 19854497; PMCID: PMC2867670.
- Sestito R, Cianfrocca R, Tocci P, Rosanò L, Sacconi A, Blandino G, Bagnato A. Targeting endothelin 1 receptor-miR-200b/c-ZEB1 circuitry blunts metastatic progression in ovarian cancer. Commun Biol. 2020 Nov 13;3(1):677. doi: 10.1038/s42003-020-01404-3. PMID: 33188287; PMCID: PMC7666224.
- Zamaraev AV, Volik PI, Sukhikh GT, Kopeina GS, Zhivotovsky B. Long non-coding RNAs: A view to kill ovarian cancer. Biochim Biophys Acta Rev Cancer. 2021 Aug;1876(1):188584. doi: 10.1016/j.bbcan.2021.188584. Epub 2021 Jun 19. PMID: 34157315.
- Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016 Mar 15;7(11):12598-611. doi: 10.18632/oncotarget.7181. PMID: 26863568; PMCID: PMC4914307.
- Schwarzenbach H, Gahan PB. Circulating non-coding RNAs in recurrent and metastatic ovarian cancer. Cancer Drug Resist. 2019 Sep 19;2(3):399-418. doi: 10.20517/cdr.2019.51. PMID: 35582568; PMCID: PMC8992516.
- Oliveira DVNP, Prahm KP, Christensen IJ, Hansen A, Høgdall CK, Høgdall EV. Noncoding RNA (ncRNA) Profile Association with Patient Outcome in Epithelial Ovarian Cancer Cases. Reprod Sci. 2021 Mar;28(3):757-765. doi: 10.1007/s43032-020-00372-7. Epub 2020 Oct 30. PMID: 33125686; PMCID: PMC7862201.
- Dianatpour A, Ghafouri-Fard S. Long Non Coding RNA Expression Intersecting Cancer and Spermatogenesis: A Systematic Review. Asian Pac J Cancer Prev. 2017 Oct 26;18(10):2601-2610. doi: 10.22034/APJCP.2017.18.10.2601. PMID: 29072050; PMCID: PMC5747377.
- Wang R, Lu X, Yu R. Lycopene Inhibits Epithelial-Mesenchymal Transition and Promotes Apoptosis in Oral Cancer via PI3K/AKT/m-TOR Signal Pathway. Drug Des Devel Ther. 2020 Jun 24;14:2461-2471. doi: 10.2147/DDDT.S251614. PMID: 32606612; PMCID: PMC7321693.
- Seyed Hosseini E, Nikkhah A, Sotudeh A, Alizadeh Zarei M, Izadpanah F, Nikzad H, Haddad Kashani H. The impact of LncRNA dysregulation on clinicopathology and survival of pancreatic cancer: a systematic review and meta-analysis (PRISMA compliant). Cancer Cell Int. 2021 Aug 23;21(1):447. doi: 10.1186/s12935-021-02125-1. PMID: 34425840; PMCID: PMC8383355.
- Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, Pourhanifeh MH, Savardashtaki A, Mirzaei H. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019 Sep 3;12(1):84. doi: 10.1186/s13048-019-0558-5. PMID: 31481095; PMCID: PMC6724287.
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015 Jan 27;5:8057. doi: 10.1038/srep08057. PMID: 25624062; PMCID: PMC4306919.
- Yang X, Mei J, Wang H, Gu D, Ding J, Liu C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020 Jun 23;20:265. doi: 10.1186/s12935-020-01367-9. PMID: 32587475; PMCID: PMC7313187.
- Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, Yang J, Shen K, Huang Y, Lang J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018 Dec;53(6):2637-2646. doi: 10.3892/ijo.2018.4566. Epub 2018 Sep 20. PMID: 30272264.
- Peng EY, Shu Y, Wu Y, Zeng F, Tan S, Deng Y, Deng Y, Chen H, Zhu L, Xu H. Presence and diagnostic value of circulating tsncRNA for ovarian tumor. Mol Cancer. 2018 Nov 22;17(1):163. doi: 10.1186/s12943-018-0910-1. PMID: 30466461; PMCID: PMC6251159.
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):57-74. doi: 10.1038/nature11247. PMID: 22955616; PMCID: PMC3439153.
- Fatima F, Nawaz M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA. 2017 Feb 23;3(1):10. doi: 10.3390/ncrna3010010. PMID: 29657282; PMCID: PMC5831998.
- Li J, Ju J, Ni B, Wang H. The emerging role of miR-506 in cancer. Oncotarget. 2016 Sep 20;7(38):62778-62788. doi: 10.18632/oncotarget.11294. PMID: 27542202; PMCID: PMC5308765.
- Luo J, Zhu C, Wang H, Yu L, Zhou J. MicroRNA-126 affects ovarian cancer cell differentiation and invasion by modulating expression of vascular endothelial growth factor. Oncol Lett. 2018 Apr;15(4):5803-5808. doi: 10.3892/ol.2018.8025. Epub 2018 Feb 12. PMID: 29552211; PMCID: PMC5840569.
- Chung S. False-positive elevations in carcinoembryonic antigen levels at a health screening center. 2019; 9:146. https://doi.org/10.3343/lmo.2019.9.3.146
- Wang HY, Hsieh CH, Wen CN, Wen YH, Chen CH, Lu JJ. Cancers Screening in an Asymptomatic Population by Using Multiple Tumour Markers. PLoS One. 2016 Jun 29;11(6):e0158285. doi: 10.1371/journal.pone.0158285. PMID: 27355357; PMCID: PMC4927114.
- Gawel SH, Jackson L, Jeanblanc N, Davis GJ. Current and future opportunities for liquid biopsy of circulating biomarkers to aid in early cancer detection. J Cancer Metastasis Treat. 2022; 8:26. http://dx.doi.org/10.20517/2394-4722.2022.13
- Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis. 2000 Mar;21(3):443-52. doi: 10.1093/carcin/21.3.443. PMID: 10688864.
- Mio C, Damante G. Challenges in promoter methylation analysis in the new era of translational oncology: a focus on liquid biopsy. Biochim Biophys Acta Mol Basis Dis. 2022 Jun 1;1868(6):166390. doi: 10.1016/j.bbadis.2022.166390. Epub 2022 Mar 14. PMID: 35296416.