Abstract
Review Article
The advances and challenges of Gene Therapy for Duchenne Muscular Dystrophy
Jacques P Tremblay* and Jean-Paul Iyombe-Engembe
Published: 25 July, 2017 | Volume 1 - Issue 1 | Pages: 019-036
Since the discovery of the dystrophin gene (DMD gene) thirty years ago, several therapeutic approaches have been investigated to treat Duchenne muscular dystrophy (DMD). This includes cell therapy, exon jumping, exonic knockout, and the CinDel method. In this article, we present the challenges of developping a treatment for DMD and the advances of these various approaches. We included the new CRISPR-Cas9 system, which permits not only major progress in the development of new treatments based on genome editing but also the production of new animal models.
Read Full Article HTML DOI: 10.29328/journal.jgmgt.1001003 Cite this Article Read Full Article PDF
Keywords:
Gene therapy, Duchenne muscular dystrophy, CRISPR/Cas9, Animal model
References
- Consortium, IHGS (2004). Nature 431: 931-945.
- Consortium, TEP (2012). Nature 489: 45-77.
- Kunkel L, Hejtmancik J, Caskey C, Speer A, Monaco AP, et al. Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature. 1986; 322: 73-77. Ref.: https://goo.gl/MtkNij
- Bodrug SE, Ray PN, Gonzalez IL, Schmickel RD, Sylvester JE, et al. Molecular analysis of à constitutional X-autosome translocation in a female with muscular dystrophy. Science.1987; 237: 1620-1624. Ref.: https://goo.gl/XC8UBR
- Hoffman EP, Brown RH Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987; 51: 919-928. Ref.: https://goo.gl/Mvt6Xz
- Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, et al. Characterization of dystrophin in muscle biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N Engl J Med.1988; 318: 1363-1368. Ref.: https://goo.gl/iACqAb
- Partridge TA, Grounds M, Sloper JC. Evidence of fusion between host and donor myoblasts in skeletal muscle grafts. Nature. 1978; 273: 306-308. Ref.: https://goo.gl/eBeWSs
- Skuk D. Myoblast transplantation for inherited myopathies: a clinical approach. Expert Opin Biol Ther. 2004; 4: 1871-1885. Ref.: https://goo.gl/g7ct6W
- Mendell JR, Campbell K, Rodino‐Klapac L, Sahenk Z, Shilling C, et al. Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010; 363: 1429-1437. Ref.: https://goo.gl/E1YG6t
- van Deutekom, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, et al. Local dystrophin restoration with antisens oligonucléotides PRO051. N Engl J Med. 2007; 357 : 2677-2686. Ref.: https://goo.gl/o9btPx
- Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo‐controlled phase 2 study. Lancet Neurol. 2014; 13: 987-996. Ref.: https://goo.gl/kJAv3X
- Gouble A, Smith J, Bruneau S, Perez C, Guyot V, et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J Gene Med. 2006; 8: 616-622. Ref.: https://goo.gl/fv4nKV
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996; 93: 1156-1160. Ref.: https://goo.gl/adR8MB
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nature Reviews Molecular Cell Biology. 2013; 14: 49-55. Ref.: https://goo.gl/azsxQ4
- Jinek M, Chvlinski K, Fonfara I, Hauer M, Doudna JA, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337: 816-821. Ref.: https://goo.gl/CmHA49
- Watt DJ, Lambert K, Morgan JE. Incorporation of donor muscle precursor cell into an area of muscle generation in the host mouse. J neurol Sci. 1982; 57: 319-331.
- Lipton BH, Schultz E. Developmental faite of skeletal muscle satellite cells. Science. 1979; 205: 1292-1294. Ref.: https://goo.gl/jy1GiZ
- Yao SN, Kurachi K. Implanted myoblasts not fuse with myofibers but also survive as muscle precursor cells. J Cell Sci. 1993; 105: 957-963. Ref.: https://goo.gl/8DDr6R
- Skuk D. Cell Transplantation and ‘‘Stem Cell Therapy’’ in the treatment of myopathies: many promises in mice, few realities in humans. Hindawi Publishing Corporation ISRN Transplantation. 2011; 25.
- Zheng B, Cao B, Crisan M, Sun B, Li G, et al. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nature Biotechnology. 2007; 25: 1025-1034. Ref.: https://goo.gl/u1gywA
- Sampaolesi M, Blot S, D’Antona G, Granger M, Tonlorenzi R, et al. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2013; 494. Ref.: https://goo.gl/z9h9Hy
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008; 3: 301-313. Ref.: https://goo.gl/FJVRMZ
- Benchaouir R, Meregalli M, Farini A, D’Antona G, Belicchi M, et al. Restoration of human dystrophin following transplantation of exon-skipping engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007; 1: 646-657. Ref.: https://goo.gl/uRsuCx
- Vauchez K, Marolleau J, Schmid M, Khattar P, Chapel A, et al. Aldehyde dehydrogenase activity identifies a population of human skeletal muscle cells with high myogenic capacities. Molecular therapy, 2009; 17: 1948-1958. Ref.: https://goo.gl/td98pa
- Mitchell KJ, Pannerec A, Cabot B, Parlakian A, Besson V, et al. Identification and characterization of à non-satellite cell muscle resident progenitor during postnatal development. Nature Cell Biology. 2010; 12: 257-266. Ref.: https://goo.gl/BjVqCZ
- Liadaki K, Casar JC, WMe. Beta-4-integrin marks interstitial myogenic progenitor cells in adult murine skeletal muscle, Journal of Histochemistry and Cytochemistry. 2012; 60: 31-44.
- Zhao C, Farruggio AP, Bjornson CRR, Chavez CL, Geisinger JM, et al. Recombinase-Mediated Reprogramming and Dystrophin Gene Addition in mdx Mouse Induced Pluripotent Stem Cells. PLoS One. 2014; 9 : e96279. Ref.: https://goo.gl/zPS8gz
- Skuk D, Goulet M, Roy B, Chapdelaine P, Bouchard JP, et al. Dystrophin expression in muscles of Duchenne muscular patients after high-density injection of normal myogenic cells. J Neuropathol Exp Neurol. 2006; 65: 371-386. Ref.: https://goo.gl/iBPfD2
- Lazerges C, Daussin PA, Coulet B, Boudbaker el Andalousi R, Micallef JP, et al. Transplantation of primary satellite cells improves properties of reinnervated skeletal muscles. Muscle Nerve. 2004; 29: 218-226. Ref.: https://goo.gl/W3VRyf
- Skuk D, Tremblay JP. Intramuscular cell transplantation as a potential treatment of myopathies: clinical and preclinical relevant data. Expert Opin Biol Ther. 2011; 11: 359-374. Ref.: https://goo.gl/3bxS1u
- Irintchev A, Langer M, Zweyer M, Theisen R, Wemig A. Functional improvement of damaged adult mouse muscle by implantation of primary myoblasts. J physiol. 1997; 1500: 775-785. Ref.: https://goo.gl/u5wJsw
- Wernig A, Zweyer M, Irintchev A. Function of skeletal muscle tissue formed after transplantation indo irradiated mouse muscles. J Physiol. 2000; 522: 333-345. Ref.: https://goo.gl/7RkSwF
- Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, et al. Human circulating AC133+ stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle, Journal of Clinical Investigation. 2004; 114: 182-195. Ref.: https://goo.gl/WQpNGc
- Hardiman O, Sklar RM, Brown HR Jr. Direct effects of cyclosporin A and cyclophosphamide on differentiation of normal human myoblasts in culture. Neurology. 1993; 43: 1432-1434. Ref.: https://goo.gl/dQY6U7
- Kinoshita I, Vilquin JT, Guerette B, Asselin I, Roy R, et al. Very efficient myoblast allotransplantation in mice under FK506 immunosuppression. Muscle & Nerve. 1994; 17: 1407-1415. Ref.: https://goo.gl/swTnwK
- Chandrasekharan D, Issa F, Wood KJ. Achieving operational tolerance in transplantation: how can lessons from the clinic inform research directions?. Transplant International. 2013; 26 : 576-589. Ref.: https://goo.gl/jWEoxX
- Arpke, Darabi R, Mader TL, Zhang Y, Toyama A, et al. A new immuno- dystrophin-deficient model, the NSG-mdx(4Cv) mouse, provides evidence for functional improvement following allogeneic satellite cell transplantation. Stem cells. 2013; 31: 1611-1620. Ref.: https://goo.gl/2SrqUy
- Fukada S, Miyagoe-Suzuki Y, Tsukihara H, Yausa K, Higuchi S, et al. Muscle regeneration by reconstitution with bone marrow or fetal liver cells from green fluorescent protein-gene transgenic mice. J Cell Sci. 2002; 115: 1285-1293. Ref.: https://goo.gl/dJ1Yn3
- Partridge TA, Morgan JE, Coulton GR, Hoffman EP, Kunkel LM. Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature. 1989; 337: 176-179. Ref.: https://goo.gl/BNw5ic
- Traggiai E, Chicha L, Mazzucchelli L, Bronz L, Piffaretti JC, et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004; 304: 104-107. Ref.: https://goo.gl/yYFWMx
- El Fahime E, Mills P, Lafreniere JF, Torrente Y, Tremblay JP. The urokinase plasminogen activator: an interesting way to improve myoblast migration following their transplantation. Exp Cell Res. 2002; 280: 169-178. Ref.: https://goo.gl/nxGa13
- Ferraro E, Cecconi F. Autophagic and apoptic response to stress signals in mammalian cells. Archives of Biochemistry and Biophysics. 2007; 462: 210-219. Ref.: https://goo.gl/QqBjpc
- Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988; 53: 219-228. Ref.: https://goo.gl/cG9dnz
- Cohen C, Parry A D. a-helical coiled coils; à widespread motif in proteins. Trends Biochem Sci. 1986; 11: 245-248.
- AE, E (1990). Dystrophin function. Lancet 335: 1289.
- Outersrout DG, Kabadi AM, Majoros WH, Reddy TE, Thakore PI, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015; 6:6244. Ref.: https://goo.gl/wRi4wk
- Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, et al. The TREAT‐NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015; 36: 395-402. Ref.: https://goo.gl/1BTyPF
- McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech. 2015; 8: 195-213. Ref.: https://goo.gl/Ric8Wq
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009; 119: 624-635. Ref.: https://goo.gl/5bvFdS
- Guiraud S, Chen H, Burns DT, Davies KE. Advances in genetic therapeutic strategies for Duchenne muscular dystrophy. Experimental Physiology. 2015; 100: 1458-1467. Ref.: https://goo.gl/329EDH
- Hoffman E. Genotype/phenotype correlations in Duchenne/Becker dystrophy. Mol Cell Biol Hum Dis Ser. 1993; 3: 12-36. Ref.: https://goo.gl/GR1aVE
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, et al. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990; 343: 180-182. Ref.: https://goo.gl/EsyThj
- Hayashita‐Kinoh H, Yugeta N, Okada H, Nitahara‐Kasahara Y, Chiyo T, et al. Intra‐amniotic rAAV‐mediated microdystrophin gene transfer improves canine X‐linked muscular dystrophy and may induce immune tolerance. Mol Ther. 2015; 23: 627-637. Ref.: https://goo.gl/ZeeCDD
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002; 8: 253-261. Ref.: https://goo.gl/Aty4cb
- Malik V, Rodino‐Klapac LR, Viollet L, Wall C, King W, et al. Gentamicin‐induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010; 67: 771-780. Ref.: https://goo.gl/KjGncc
- Aartsma-Rus A, van Ommen GJ. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA. 2007; 13: 1609-1624. Ref.: https://goo.gl/THGt6o
- Dunkley MG, Manoharan M, Villiet P, Eperon IC, Dickson G. Modification of splicing in the dystrophin gene in cultured mdx muscle cells by antisenses oligoribonucleotides. Hum mol Genet. 1998; 7: 1083-1090. Ref.: https://goo.gl/KB9fSM
- van Deutekom JC, Bremmer‐Bout M, Janson AA, Ginjaar IB, Baas F, et al. Antisense‐induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet. 2001; 10: 1547-1554. Ref.: https://goo.gl/gJUPGS
- Kinali M, Arechavala‐Gomeza V, Feng L, Cirak S, Hunt D, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI‐4658 in Duchenne muscular dystrophy: a single‐blind, placebo‐controlled, dose‐escalation, proof‐of‐concept study. Lancet Neurol. 2009; 8: 918-928. Ref.: https://goo.gl/rMgxds
- Mendell JR, Rodino‐Klapac LR, Sahenk Z, Roush K, Bird L, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013; 74: 637-647. Ref.: https://goo.gl/TkUMi8
- http://www.gsk.com
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, et al. Rescue of dystrophic muscle through U7 snRNA‐mediated exon skipping. Science. 2004; 306: 1796-1799. Ref.: https://goo.gl/WBj2PP
- Goyenvalle A, Griffith G, Babbs A, EI Andaloussi S, Ezzat K, et al. Functional correction in mouse models of muscular dystrophy using exon‐skipping tricyclo‐DNA oligomers. Nat Med. 2015; 21: 270-275. Ref.: https://goo.gl/wT3Dbe
- http://www.streetinsider.com/...Sarepta+Therapeutics+(SRPT)+Eteplirsen
- Béroud C, Tuffery‐Giraud S, Matsuo M, Hamroun D, Humbertclaude V, et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat. 2007; 28: 196-202. Ref.: https://goo.gl/1burBb
- Stamler JS, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001; 81: 209-237. Ref.: https://goo.gl/LtpbR8
- Bredt DS. Endogenous nitric oxide synthesis: biological functions and pathophysiology. Free Radic Res. 1999; 31: 577-596. Ref.: https://goo.gl/sZN9wb
- Heydemann, AaM E. NO more muscle fatigue. J Clin Invest. 2009; 119: 448-450.
- Jinek M, Chylinski K, Fonfara I, Hauer JA, Charpentier E. A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337: 816-821. Ref.: https://goo.gl/rwQsCi
- Jarmin S, Kymalainen H, Popplewell L, Dickson G. New development in the use of gene therapy to treat Duchenne muscular dystrophy. Expert Opin Biol Ther. 2014; 14: 209-230. Ref.: https://goo.gl/VaJhWC
- Popplewell L, Koo T, Leclerc X, Duclert A, Mamchaoui K, et al. Gene correction of a duchenne muscular dystrophy mutation by meganuclease-enhanced exon knock-in. Hum Gene Ther. 2013; 24: 692-701. Ref.: https://goo.gl/c9sTMY
- Ousterout D, Kabadi AM, Thakore PI, Perez-Pinera P, Brown M, et al. Correction of Dystrophin Expression in Cells From Duchenne Muscular Dystrophy Patients Through Genomic Excision of Exon 51 by Zinc Finger Nucleases. Mol Ther. 2015; 23: 523-532. Ref.: https://goo.gl/MzTxtF
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cell by TALENs and CRISPR-Cas9. Stem Cell Reports. 2015; 4: 134-154. Ref.: https://goo.gl/DpDtH7
- Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, et al. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004; 43: 14015-14026. Ref.: https://goo.gl/FdVeL1
- Grizot S, Epinat JC, Thomas S, Duclert A, Rolland S, et al. Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds. Nucleic Acids Res. 2010; 38: 2006-2018. Ref.: https://goo.gl/6NFHwf
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011; 11: 11-27. Ref.: https://goo.gl/vM98zr
- Chapdelaine P, Pichavant C, Rousseau J, Pâques F, Tremblay JP. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 2010; 17: 846-858. Ref.: https://goo.gl/QV9mrz
- Klug A. The discovery of Zinc Fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010; 79: 213-231. Ref.: https://goo.gl/MSdK7H
- Kim YG, Li L, Chandrasegaran S. Insertion and deletion mutants of FokI restriction endonuclease. J Biol Chem. 1994; 269: 31978-31982. Ref.: https://goo.gl/cixgTn
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011; 8: 74-79. Ref.: https://goo.gl/stXgXs
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007; 25: 778-785. Ref.: https://goo.gl/i6fbF8
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007; 25: 786-793. Ref.: https://goo.gl/z4UxSJ
- Anguela XM, Sharma R, Doyon Y, Miller JC, Li H, et al. Robust ZFN-mediated genome editing in adule hemophilic mice. Blood.2013; 122: 3283-3287. Ref.: https://goo.gl/NZvMJe
- Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol.2012; 23: 1-7. Ref.: https://goo.gl/ajCo7C
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009; 326: 1509-1512. Ref.: https://goo.gl/etRHCy
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009; 326: 1501. Ref.: https://goo.gl/iZLHkR
- Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010; 13: 394-401. Ref.: https://goo.gl/EpYj2E
- Hisano Y, Ota S, Arakawa K, Muraki M, Kono N, et al. Quantitative assay for TALEN activity at endogenous genomic loci. Biol Open. 2013; 2: 363-367. Ref.: https://goo.gl/VEi2Ev
- Ousterout DG, Perez-Pinera P, Thakore PI, Kabadi AM, Brown MT, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther. 2013; 21: 1718-1726. Ref.: https://goo.gl/my1vk1
- Jinek M, East A, Cheng A, Lin S, Ma E, et al. RNA-programmed genome editing in human Cells. ELife. 2013; 2. Ref.: https://goo.gl/a964dZ
- Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013; 339: 819-823. Ref.: https://goo.gl/gQMiSy
- Doudna J, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346: Ref.: https://goo.gl/Pg9LmR
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014; 32: 347-355. Ref.: https://goo.gl/26DN9d
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013; 339: 823-826. Ref.: https://goo.gl/1nBQCh
- Cho S, Kim S, Kim JK, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013; 31: 230-232. Ref.: https://goo.gl/uSPnNX
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015; 163: 1-13. Ref.: https://goo.gl/SJmEB3
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520: 186-191. Ref.: https://goo.gl/t3uR9f
- Kim E, Koo T, Park SW, Kim D, Kim K, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017; 8: Ref.: https://goo.gl/osgJzU
- Zheng Q, Cai X, Tan M, Schaffert S, Arnold C, et al. Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques. 2014; 57: 115-124. Ref.: https://goo.gl/LgZzML
- Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017; 8: Ref.: https://goo.gl/VbqZeq
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2015; 35: 407-411. Ref.: https://goo.gl/qLqCG8
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, et al. In vivo genome editing imporves muscle function in a model of Duchenne muscular dystrophy. Science.2015; 350: 403-407. Ref.: https://goo.gl/8z98NS
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016; 351: 400-403. Ref.: https://goo.gl/DpbqgY
- Nicolas A, Raguénès-Nicol C, Ben Yaou R, Ameziane-Le Hir S, Chéron A. Becker muscular dystrophy severity is linked to the structure of dystrophin. Hum Mol Genet. 2015; 24: 1267-1279. Ref.: https://goo.gl/vgcWbq
- Iyombe-Engembe JP, Ouellet DL, Barbeau X, Rousseau J, Chapdelaine P, et al. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol Ther Nucleic Acids. 2016; 5: e283. Ref.: https://goo.gl/dzBUqc
- Tremblay JP, Iyombe-Engembe JP, Duchêne B, Ouellet DL. Gene Editing for Duchenne Muscular Dystrophy Using the CRISPR/Cas9 Technology: The Importance of Fine-tuning the Approach. Mol Ther. 2016; 24: 1888-1889. Ref.: https://goo.gl/5qTD6i
- Farboud B, Meyer BJ. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015; 199: 959-971. Ref.: https://goo.gl/qPtn3i
- http://www.askbio.comm
- http://www.ptcbio.com
- Bushby K, Finkel R, Wong B, Barohn R, Campbell C, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve.2014; 50: 477-487. Ref.: https://goo.gl/96tuKV
- Bulfield C, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984; 1: 1189-1192. Ref.: https://goo.gl/LFz1gt
- Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992; 360: 588-591. Ref.: https://goo.gl/xSyP6r
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell.2010; 143: 1059-1071. Ref.: https://goo.gl/Li97aB
- Chandrasekharan K, Yoon JH, Xu Y, deVries S, Camboni M, et al. A human-specific deletion in mouse Cmah increases disease severity in the mdx model of Duchenne muscular dystrophy. Sci Transl Med. 2010; 2: Ref.: https://goo.gl/ksFNWv
- Bodor M, McDonald CM. Why short stature is beneficial in Duchenne muscular dystrophy. Muscle Nerve. 2013; 48: 336-342. Ref.: https://goo.gl/vQFJpG
- Hoen PA, de Meijer EJ, Boer JM, Vossen RH, Turk R, et al. Generation and Characterization of Transgenic Mice with the Full-length Human DMD Gene. J Biol Chem. 2008; 283: 5899-5907. Ref.: https://goo.gl/yELG4v
- INNES JR. Myopathies in animals; a record of some cases including progressive muscular dystrophy (pseudo-hypertrophic) (dog), “weisses Fleisch” (lamb), neuropathic muscular atrophy (sheep) and lymphocytic/histiocytic myositis, neuritis, radiculitis (dog). Br Vet J. 1951; 107: 131-143. Ref.: https://goo.gl/UXKGKh
- Sharp NJ, Kornegay JN, Van Camp SD, Herbstreith MH, Secore SL, et al. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics. 1992; 13: 115-121. Ref.: https://goo.gl/QCSR8f
- Cooper BJ, Valentine BA, Wilson S, Patterson DF, Concannon PW. Canine muscular dystrophy: confirmation of X-linked inheritance. J Hered. 1988; 79: 405-408. Ref.: https://goo.gl/rPTEK1
- Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, et al. A Duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient Cavalier King Charles spaniels is amenable to exon 51 skipping. PLoS One. 2010; 5: e8647. Ref.: https://goo.gl/iwbE3Y
- Nakamura K, Fuji W, Tsuboi M, Tanihata J, Teramoto N, et al. Generation of muscular dystrophy rat with CRISPR/Cas system. Sci Rep. 2014; 4: Ref.: https://goo.gl/KZNHY8
- Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, et al. Characterization of dystrophin deficient rats : a new model for Duchenne muscular dystrophy. Plos One. 2014; 9: e110371. Ref.: https://goo.gl/kMuk87
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet.2015; 24: 3764-3774. Ref.: https://goo.gl/y7uuSG
- Klymiuk N, Blutke A, Graf A, Krause S, Burkhardt K, et al. Dystrophin-deficient pigs provide new insights into hierarchy of physiology derangements of dystrophic muscle. Hum Mol Genet. 2013;22: 4368-4382. Ref.: https://goo.gl/LnqtKi
Figures:
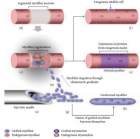
Figure 1
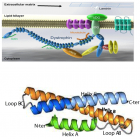
Figure 2
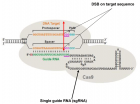
Figure 3
Similar Articles
Recently Viewed
-
Anxiety and depression as an effect of birth order or being an only child: Results of an internet survey in Poland and GermanyJochen Hardt*,Lisa Weyer,Malgorzata Dragan,Wilfried Laubach. Anxiety and depression as an effect of birth order or being an only child: Results of an internet survey in Poland and Germany. Insights Depress Anxiety. 2017: doi: 10.29328/journal.hda.1001003; 1: 015-022
-
Transumbilical Single-incision Hiatal Hernia Repair and Nissen Fundoplication in situs Inversus Totalis: A Rare Case ReportQing Cao,Chen Kang,Kang Gu,Yin Peng,Yang Lv,Xu-Zhong Ding,Peng Li*. Transumbilical Single-incision Hiatal Hernia Repair and Nissen Fundoplication in situs Inversus Totalis: A Rare Case Report. Adv Treat ENT Disord. 2026: doi: 10.29328/journal.ated.1001017; 10: 001-003
-
Case Report of a Child with Beta Thalassemia Major in a Tribal Region of IndiaNeha Chauhan, Prakash Narayan, Mahesh Narayan, Manisha Shukla*. Case Report of a Child with Beta Thalassemia Major in a Tribal Region of India. J Child Adult Vaccines Immunol. 2023: doi: 10.29328/journal.jcavi.1001011; 7: 005-007
-
Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging PresentationKarthik Baburaj*, Priya Thottiyil Nair, Abeed Hussain, Vimal MV. Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging Presentation. Ann Clin Endocrinol Metabol. 2024: doi: 10.29328/journal.acem.1001029; 8: 001-003
-
Physical Performance in the Overweight/Obesity Children Evaluation and RehabilitationCristina Popescu, Mircea-Sebastian Șerbănescu, Gigi Calin*, Magdalena Rodica Trăistaru. Physical Performance in the Overweight/Obesity Children Evaluation and Rehabilitation. Ann Clin Endocrinol Metabol. 2024: doi: 10.29328/journal.acem.1001030; 8: 004-012
Most Viewed
-
Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization)Dasse Sery Romuald*, KL Siransy, N Koffi, RO Yeboah, EK Nguessan, HA Adou, VP Goran-Kouacou, AU Assi, JY Seri, S Moussa, D Oura, CL Memel, H Koya, E Atoukoula. Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization). Arch Asthma Allergy Immunol. 2024 doi: 10.29328/journal.aaai.1001035; 8: 007-012
-
Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian RandomizationYong-Qing Zhu, Xiao-Yan Meng, Jing-Hua Yang*. Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian Randomization. Arch Asthma Allergy Immunol. 2023 doi: 10.29328/journal.aaai.1001032; 7: 012-022
-
An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergiesNathalie Cottel,Aïcha Dieme,Véronique Orcel,Yannick Chantran,Mélisande Bourgoin-Heck,Jocelyne Just. An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergies. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001027; 5: 030-037
-
Snow white: an allergic girl?Oreste Vittore Brenna*. Snow white: an allergic girl?. Arch Asthma Allergy Immunol. 2022 doi: 10.29328/journal.aaai.1001029; 6: 001-002
-
Cytokine intoxication as a model of cell apoptosis and predict of schizophrenia - like affective disordersElena Viktorovna Drozdova*. Cytokine intoxication as a model of cell apoptosis and predict of schizophrenia - like affective disorders. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001028; 5: 038-040

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."