Figure 3
Reasons why new coronavirus, SARS-CoV-2 infections are likely to spread
Takuma Hayashi*, Takashi Ura, Kaoru Abiko, Masaki Mandan, Nobuo Yaegashi and Ikuo Konishi
Published: 28 April, 2020 | Volume 3 - Issue 1 | Pages: 001-003
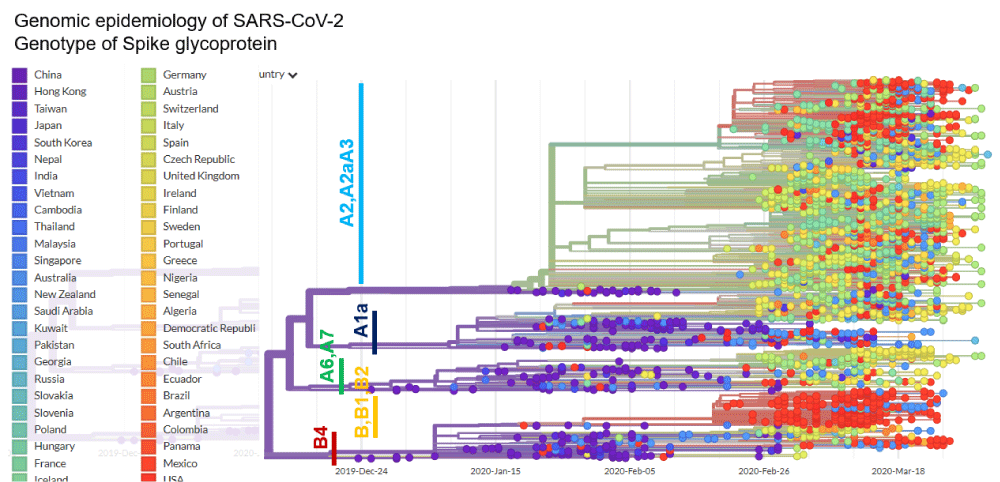
Figure 3:
Phylogenetic network analysis of amino acid sequences of spike glycoprotein derived from 160 complete human SARS-Cov-2 genomes. Our research results find ten central variants distinguished by amino acid changes, which we have named A1a, A2, A2a, A3, A6, A7, B, B1, B2, and B4, with being the ancestral type according to the bat out group coronavirus. The A1a, A6, A7 and B4 types are found in significant proportions in East Asia, A2, A2a, and A3 types are found in Europeans, B, B1, and B2 types are found in Americans. In contrast, the B4 type is the most common type in America and East Asia.
Read Full Article HTML DOI: 10.29328/journal.jgmgt.1001005 Cite this Article Read Full Article PDF
More Images
Similar Articles
-
Reasons why new coronavirus, SARS-CoV-2 infections are likely to spreadTakuma Hayashi*,Takashi Ura,Kaoru Abiko,Masaki Mandan,Nobuo Yaegashi,Ikuo Konishi. Reasons why new coronavirus, SARS-CoV-2 infections are likely to spread. . 2020 doi: 10.29328/journal.jgmgt.1001005; 3: 001-003
Recently Viewed
-
Knowledge, Attitude, and Practice of Healthcare Workers in Ekiti State, Nigeria on Prevention of Cervical CancerAde-Ojo Idowu Pius*, Okunola Temitope Omoladun, Olaogun Dominic Oluwole. Knowledge, Attitude, and Practice of Healthcare Workers in Ekiti State, Nigeria on Prevention of Cervical Cancer. Arch Cancer Sci Ther. 2024: doi: 10.29328/journal.acst.1001038; 8: 001-006
-
Oral Cancer Management is not just Treatment! But also, how early Pre-cancerous Lesions are Diagnosed & Treated!!Suresh Kishanrao*. Oral Cancer Management is not just Treatment! But also, how early Pre-cancerous Lesions are Diagnosed & Treated!!. Arch Cancer Sci Ther. 2024: doi: 10.29328/journal.acst.1001039; 8: 007-012
-
Breast Cancer in FemaleLorena Menditto*. Breast Cancer in Female. Arch Cancer Sci Ther. 2024: doi: 10.29328/journal.acst.1001040; 8: 013-018
-
Lived Experiences of Cervical Cancer Patients Receiving Chemotherapy at Cancer Diseases Hospital in Lusaka, ZambiaElisha Benkeni Kapya*, Marjorie Kabinga Makukula, Mwaba Chileshe Siwale, Victoria Kalusopa Mwiinga, Elijah Mpundu. Lived Experiences of Cervical Cancer Patients Receiving Chemotherapy at Cancer Diseases Hospital in Lusaka, Zambia. Arch Cancer Sci Ther. 2024: doi: 10.29328/journal.acst.1001041; 8: 019-036
-
Accessory Splenic Mass Masquerading as Hepatocellular Carcinoma: A Diagnostic DilemmaSoe P Winn*, Tharun Shyam, M Isabel Fiel, Yiwu Huang. Accessory Splenic Mass Masquerading as Hepatocellular Carcinoma: A Diagnostic Dilemma. Arch Cancer Sci Ther. 2024: doi: 10.29328/journal.acst.1001042; 8: 037-040
Most Viewed
-
Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trialSathit Niramitmahapanya*,Preeyapat Chattieng,Tiersidh Nasomphan,Korbtham Sathirakul. Effects of dietary supplementation on progression to type 2 diabetes in subjects with prediabetes: a single center randomized double-blind placebo-controlled trial. Ann Clin Endocrinol Metabol. 2023 doi: 10.29328/journal.acem.1001026; 7: 00-007
-
Physical Performance in the Overweight/Obesity Children Evaluation and RehabilitationCristina Popescu, Mircea-Sebastian Șerbănescu, Gigi Calin*, Magdalena Rodica Trăistaru. Physical Performance in the Overweight/Obesity Children Evaluation and Rehabilitation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001030; 8: 004-012
-
Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging PresentationKarthik Baburaj*, Priya Thottiyil Nair, Abeed Hussain, Vimal MV. Hypercalcaemic Crisis Associated with Hyperthyroidism: A Rare and Challenging Presentation. Ann Clin Endocrinol Metabol. 2024 doi: 10.29328/journal.acem.1001029; 8: 001-003
-
Exceptional cancer responders: A zone-to-goDaniel Gandia,Cecilia Suárez*. Exceptional cancer responders: A zone-to-go. Arch Cancer Sci Ther. 2023 doi: 10.29328/journal.acst.1001033; 7: 001-002
-
The benefits of biochemical bone markersSek Aksaranugraha*. The benefits of biochemical bone markers. Int J Bone Marrow Res. 2020 doi: 10.29328/journal.ijbmr.1001013; 3: 027-031

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."